 |
 |
| Plant Pathol J > Volume 34(4); 2018 > Article |
Abstract
Northern corn leaf spot and southern corn leaf blight caused by Cochliobolus carbonum (anamorph, Bipolaris zeicola) and Cochliobolus heterostrophus (anamorph, Bipolaris maydis), respectively, are common maize diseases in Korea. Accurate detection of plant pathogens is necessary for effective disease management. Based on the polyketide synthase gene (PKS) of Cochliobolus carbonum and the nonribosomal peptide synthetase gene (NRPS) of Cochliobolus heterostrophus, primer pairs were designed for PCR to simultaneously detect the two fungal pathogens and were specific and sensitive enough to be used for duplex PCR analysis. This duplex PCR-based method was found to be effective for diagnosing simultaneous infections from the two Cochliobolus species that display similar morphological and mycological characteristics. With this method, it is possible to prevent infections in maize by detecting infected seeds or maize and discarding them. Besides saving time and effort, early diagnosis can help to prevent infections, establish comprehensive management systems, and secure healthy seeds.
Maize (Zea mays L.) is one of the most widely grown crops. Maize was cultivated on approximately 15,356 ha in 2015, primarily in Gangwon province, accounting for 38% of the overall maize production area (Kang, 2013; Kim, 2017). A significant limitation to obtaining good maize yield is a fungal infection. Thirteen fungal species of maize have been reported in Korea (KSPP, 2009; Shin, 2014), and recently, the incidences of infections with northern corn leaf spot (NCLS) and southern corn leaf blight (SLB) have increased. Northern corn leaf spot, caused by Cochliobolus carbonum (anamorph, Bipolaris zeicola), is of significant concern (Nelson, 1970; Shoemaker, 1959) because it results in the loss of crops if it develops before or during the tasseling and silking phases of maize development. Southern corn leaf blight, caused by Cochliobolus heterostrophus (anamorph, Bipolaris maydis) was a severe crop epidemic in the US in 1970 and is now primarily prevalent in tropical and subtropical maize-growing areas (Ali et al., 2011; Wang et al., 2017). This pathogen spreads from leaf litter and can produce wind-borne spores within days (Horwitz et al., 2013). NCLS and SLB are caused by pathogens of the same genus of Cochliobolus, which have similar morphological and mycological characteristics. Therefore, they are often difficult to differentiate and their accurate diagnosis remains problematic. The accurate detection and identification of the two pathogens are necessary for disease management to sustain a high yield potential of crops. The utility of PCR as a specific and sensitive assay for plant pathogen identification is well documented; regular PCR, nested PCR, multiplex PCR, and LAMP (Feng et al., 2012; Grote et al., 2002; Henson and French, 1993; Wang et al., 2010). A PCR-based detection method has been adopted as the most reliable and rapid technique (Hameed et al., 2014), among which, multiplex PCR has been developed to simultaneously detect several pathogens (Asano et al., 2010; James et al., 2006), reliably, rapidly and inexpensively.
In this study, we have established the detection method to simultaneously diagnose two fungal pathogens of the same genus. Moreover it has been confirmed that this method can be used to directly detect from maize infected with two fungal pathogens without isolating each.
The fungal strains used in this study are listed in Table 2. All fungal pathogens were isolated from upland fields and maintained in the collection of the Crop Cultivation and Environment Division of National Institute of Crop Science, including the targeted isolates Cochliobolus carbonum, and Cochliobolus heterostrophus collected from Gangwon province in Korea. To test of primer specificity, strains reported as maize fungal pathogens and saprophytes found in the soil or plant debris where maize was mainly grown were selected. Diseased tissues of maize were surface-sterilized with 70% ethyl alcohol for 1 min, washed two times with sterile water and placed it on water agar for incubation 2 days. Purely isolated pathogens were cultured in potato dextrose agar (PDA). The identification of all isolates was confirmed by conventional morphological and molecular methods. In order to observe conidia, living cultures were grown on PDA, incubated 25°C for 4 weeks, and exposed to near ultraviolet light source on a 12-h light/dark diurnal cycle (Sivanesan, 1987). Conidia were mounted on glass slides in D.W. and images captured with Olympus camera attached to the Olympus BX51 compound microscope. For molecular analysis, the primers used for universal fungal amplification were ITS4 (TCCTCCGCTTATTGTATGC) and ITS5 (GGAAGTAAAAGTCGTAACAAGG) obtained from White et al. The neighbor-joining analyses were performed using MEGA5 (Tamura et al., 2011). Phylogenetic analyses using with the ITS region were performed to identify the genetic difference between Cochliobolus carbonum F170 and Cochliobolus heterostrophus F169. These sequences were aligned with selected sequences of Cochliobolus (Bipolaris) species obtained from National Center for Biotechnology Information.
Pure cultures of Cochliobolus carbonum and Cochliobolus heterostrophus isolates were inoculated on rice bran medium (rice bran, 20 g; sugar, 20 g; agar, 20 g; water, 1 l) and incubated at 25°C for 20 days. Induction of sporulation was maximized with 12 h near ultraviolet light/12 h dark diurnal cycle at 25°C in 10 days. Following induction, the spores of each suspensions were adjusted from 105 to 106 spores/ml. The fungal spore suspensions were inoculated by spraying onto maize seedlings at the 3 leaf formation stage based on spore concentration. The symptoms were observed 4 days after inoculation.
Fungal strains were grown on potato dextrose agar (PDA) medium at 25°C. Mycelia were scraped off the PDA cultures and macerated with 0.5 mm glass beads in 1.5 ml tube. The genomic DNA of each fungus was extracted using a NucleoSpin Plant Π Kit (MACHEREY-NAGEL, Germany) as per the manufacturer’s protocol. The concentrations of the extracted DNA were determined by measuring A260/280 using a Biodrop spectrophotometer (BioDrop, UK). To direct detect in plant leaves artificially inoculated fungal pathogens, 0.5 g plant leaves were collected from each samples and ground in the presence of liquid nitrogen using mortar and pestle. The genomic DNA of each leaves was extracted using the same kit.
The Duplex PCR assay were performed using Accupower PCR premix (Bioneer, Korea) as per the manufacturer’s protocol. The final volume of the duplex PCR mix was adjusted to 20 μl with sterile distilled water, and the reaction mixture contained 1 μl (10 pmole) of specific primers for each pathogen, 1 μl (50 ng) of diluted template genomic DNA, 1 unit of Top polymerase, 250 μM dNTP mixture, 10 mM Tris-HCl (pH 9.0), 30 mM KCl. The cycling parameters included an initial denaturation step at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. PCR products were visualized on a 1.5% tris-borate-EDTA agarose gel with loading star (Dyne Bio). For direct duplex PCR, the maize leaves inoculated with each pathogens were cut in small pieces and homogenized in D.W. using Grinding Tube & Pestle (Inclon, Korea). The direct duplex PCR assay were performed using Accupower PCR premix (Bioneer, Korea) as per the manufacturer’s protocol. The final volume of the duplex PCR mix was adjusted to 20 μl with sterile distilled water, and the reaction mixture contained 1 μl (10 pmole) of specific primers for each pathogen, 3 μl of the crude supernatant sap, 1 unit of Top polymerase, 250 μM dNTP mixture, 10 mM Tris-HCl (pH 9.0), 30 mM KCl. PCR condition of direct duplex PCR is the same as that of duplex PCR assay.
To determine the detection limit of PCR, genomic DNA of Cochliobolus carbonum and Cochliobolus heterostrophus were serially diluted with distilled water to yield a final concentration ranging from 5 ng/μl to 0.05 ng/μl. The final volume of PCR mix was adjusted to 20 μl with sterile distilled water, and the reaction mixture contained 1 μl (10 pmole) of specific primers for each pathogen, 2 μl (according to each concentration) of diluted template genomic DNA, 1 unit of Top polymerase, 250 μM dNTP mixture, 10 mM Tris-HCl (pH 9.0), 30 mM KCl and enzyme mix. The cycling parameters included an initial denaturation step at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. PCR products were visualized on a 1.5% tris-borate-EDTA agarose gel with loading star (Dyne Bio).
The features of the Cochliobolus heterostrophus and Cochliobolus carbonum found in the maize field were very similar and difficult to distinguish (Fig. 1). Conidia produced by Cochliobolus heterostrophus F169 were the spindle-shaped, dark brown color and Conidia produced by Cochliobolus carbonum F170 were straight to moderately curved, occasionally cylindrical but usually broad in the middle and tapering towards the rounded ends. Conidia of Cochliobolus heterostrophus F169 was different from conidia of Cochliobolus carbonum F170 and Cochliobolus carbonum F170 and Cochliobolus heterostrophus F-169 could be distinguished in the phylogenetic tree constructed using ITS sequences (Fig. 2).
Primer pairs were designed for PCR based on the polyketide synthase (PKS) gene of Cochliobolus carbonum and the nonribosomal peptide synthetase (NRPS) gene of Cochliobolus heterostrophus (Table 1). To ensure simultaneous detection, primers were designed to produce two differently sized PCR products representing each fungal species and to use the same optimum annealing temperature, 60°C, for PCR. To confirm the correct sizes of the PCR products, duplex PCR analysis was performed for the two fungal pathogens (Fig. 3A). Duplex PCR using the four primers designed in this study amplified the respective expected targets for Cochliobolus carbonum and Cochliobolus heterostrophus (Fig. 3A), producing amplification products of different sizes for the two pathogens: Cochliobolus carbonum (268 bp) and Cochliobolus heterostrophus (539 bp). Therefore, these primer sets can be used to detect fungi of the same genus, Cochliobolus carbonum and Cochliobolus heterostrophus, in maize. To test the specificity of these primer sets, PCR amplification, using primer sets shown in Table 1, was performed with DNA from 13 different fungi (Table 2), none of which yielded the appropriate products on duplex PCR (Fig. 3B). The two sets of primers (Bm-F/R, and Bz-F/R) yielded products of 268 bp for Cochliobolus carbonum and 539 bp for Cochliobolus heterostrophus in only target strains, Cochliobolus heterostrophus F169, F217, F218 and Cochliobolus carbonum F170, F174.
To determine the sensitivity of the pathogen-specific primers, each primer set was tested with serial dilutions of the corresponding fungal genomic DNA. Fig. 4 shows a representative serial dilution of the PCR amplicons with primers Bm-F, Bm-R, and Bz-F, Bz-R. The sensitivity of each primer set ranged from 0.05-5 ng/μl, which indicated the potential to amplify the appropriate amplicon from purified genomic DNA down to 0.05 ng/μl.
The symptoms of the infections caused by the two pathogens were very similar: spindle-shaped with yellow-green halos (Fig. 1). However, the spots caused by Cochliobolus heterostrophus were larger and, fewer than those spots caused by Cochliobolus carbonum. Cochliobolus heterostrophus showed higher pathogenicity than Cochliobolus carbonum in corn seedlings following inoculation with conidial suspensions (Fig. 5B). To directly detect fungal pathogens in artificially infected maize, maize seedlings infected with Cochliobolus carbonum, Cochliobolus heterostrophus, and both pathogens were examined after directly extracting DNA without pure isolation of each pathogens. A total 12 maize leaves were tested, and amplified products of the expected sizes were identified in samples infected with Cochliobolus heterostrophus only (Fig. 5; Lanes 5-7) Cochliobolus carbonum only (Fig. 5; Lanes 8-10), and both pathogens (Fig. 5; Lanes 11-13) except in asymptomatic samples (Fig. 5; Lanes 2-4).
Cochliobolus carbonum and Cochliobolus heterostrophus cause two fungal diseases in maize, and the seeds infected by these pathogens serve as the primary infectious agents (Shurtleff, 1980). Therefore, it is necessary to periodically check whether the maize itself is infected in order to effectively block the fungal disease and to secure healthy seeds. It is difficult to determine whether infections are caused by one of them, without interference, because the two fungi share genetic characteristics. Symptoms of the infections caused by Cochliobolus heterostrophus and Cochliobolus carbonum are very similar and difficult to distinguish on the field. They can be distinguished by morphological characterization of the size of their conidia after isolation. Conidia of Cochliobolus heterostrophus are distinctly curved, whereas those of Cochliobolus carbonum are shorter and slightly curved. However, morphological differentiation of these two fungal pathogens is a cumbersome and time-consuming task because morphological and biochemical characterizations of either fungus requires pure isolation of each pathogens in separate cultures. Compared with traditional or conventional methods such as microscopic examination and pathogen isolation, PCR-based methods have the advantages of being rapid, simple, and reliable. The internal transcribed spacer (ITS) locus used in this study is the universal barcode marker for fungi (Schoch et al., 2012), and some studies have used only ITS locus to identify and describe Cochilobolus species (Ahmadpour et al., 2012; Da Cunha et al., 2012). The ITS alignment used in this study was able to differentiate Cochliobolus carbonum, and Cochliobolus heterostrophus from 13 Cochliobolus species. However, most recently, it was strongly suggested that secondary loci, such as ITS, GAPDH, TEF1α, or RPB2 should be considered together when differentiating Cochliobolus species (Berbee et al., 1999; Manamgoda et al., 2012; Tan et al., 2016). Therefore, polymerase chain reaction alone, using ITS may not reveal the distinct differences of Cochliobolus species. In addition, PCR based on secondary loci, such as ITS, GAPDH, TEF1α, or RPB2 also requires the isolation and cultivation of each fungus, and therefore, is not completely appropriate for rapid identification of plant pathogens. Our report on the use of duplex PCR for identification of Cochliobolus carbonum, and Cochliobolus heterostrophus has offered an alternative to conventional detection methods. Moreover, the method developed in this study can be used for the simultaneous diagnosis of infections caused by the two Cochliobolus species (Fig. 1). This method can also be applied to detect pathogens in the event where maize is contaminated with many different pathogens because the primer pair used in this study specifically reacted with the target pathogens causing northern corn leaf spot (NCLS) and southern corn leaf blight (SLB), but did not react with other pathogens belonging to the genus Fusarium or Rhizoctonia. Therefore, we suggest that it is more effective and rapid to use the primer pairs developed in this study rather than use some of the aforementioned loci (ITS, GAPDH, TEF1α, or RPB2) for the molecular detection of these pathogens.
In this study, the sensitivity of the duplex PCR was comparable to that of single-primer-set PCR. Since each primer pair requires different annealing temperatures and durations, it was essential to determine the optimal temperature and time conditions, so that the primers would react properly. Furthermore, with this method, the two pathogens could be analyzed simultaneously because the sizes of the products obtained with each primer pair were distinct.
Maize is one of the most important food crops with the highest production worldwide, and its economic value is also quite high. Maize diseases directly affect the production of corn, potentially leading to great economic losses. However, prompt diagnosis can save time and effort, potentially helping in the prevention of infections, establishment of comprehensive management systems, and securing of healthy seeds. This is first report on the application of the duplex PCR technique to simultaneously detect two species of the same genus, Cochliobolus carbonum and Cochliobolus heterostrophus. This technique is also specific and convenient for the direct detection of pathogens in infected maize leaves without a pure culture. It is expected to be applied for the detection of pathogens in maize cultivation fields, where rapid and accurate diagnoses of infections are required.
Acknowledgements
This study was supported by the “Cooperative Research Program for Agriculture Science & Technology Development (Project title: Field ecology and control techniques for corn smut in maize fields, Project No. PJ01182402)” Rural Development Administration, Republic of Korea.
Fig. 1
Symptoms of two fungal diseases in maize. (A) Southern corn leaf blight caused by Cochliobolus heterostrophus. (B) Northern corn leaf spot caused by Cochliobolus carbonum. (C) A microscopic image of conidia produced by Cochliobolus heterostrophus F169. (D) A microscopic image of conidia produced by Cochliobolus carbonum F170. Scale bar = 50 μm.

Fig. 2
Phylogenetic tree derived from ITS sequence of Cochliobolus sp. including the targeted strains Cochliobolus carbonum F170 and Cochliobolus heterostrophus F169. The tree was constructed using the neighbor-joining method. Bar, 0.5% estimated sequence divergence.
Fig. 3
Detection sensitivity for Cochliobolus carbonum F170 and Cochliobolus heterostrophus F169 using duplex PCR with two sets of specific primers. (A) M: molecular size marker (100-2000 bp), Lanes 1-2: Positive control (Eukaryotic translation elongation factor 1 alpha gene); Lane 3: No treatment; Lanes 4-5: Cochliobolus heterostrophus F169; Lanes 6-7: Cochliobolus carbonum F170; Lanes 8-9: Cochliobolus heterostrophus F169 + Cochliobolus carbonum F170. (B) M: molecular size marker (100-2000 bp), Lane 1: Cochliobolus heterostrophus strain F169; Lane 2: Cochliobolus heterostrophus strain F217; Lane 3: Cochliobolus heterostrophus strain F218; Lane 4: Cochliobolus carbonum strain F170; Lane 5: Cochliobolus carbonum strain F174; Lane 6: Fusarium equiseti strain F159; Lane 7: Fusarium proliferatum strain; Lane 8: Fusarium oxysporum strain; Lane 9: Fusarium solani strain; Lane 10: Fusarium graminearum stain F163; Lane 11: Fusarium verticillioides strain F157; Lane 12: Colletotrichum gloeosporioides strain; Lane 13: Myrothecium sp. strain; Lane 14: Gibberella moniliformis strain; Lane 15: Trichothecium roseum strain; Lane 16: Curvularia lunata strain; Lane 17: Phoma sp.; Lane 18: Rhizoctonia solani strain F167.
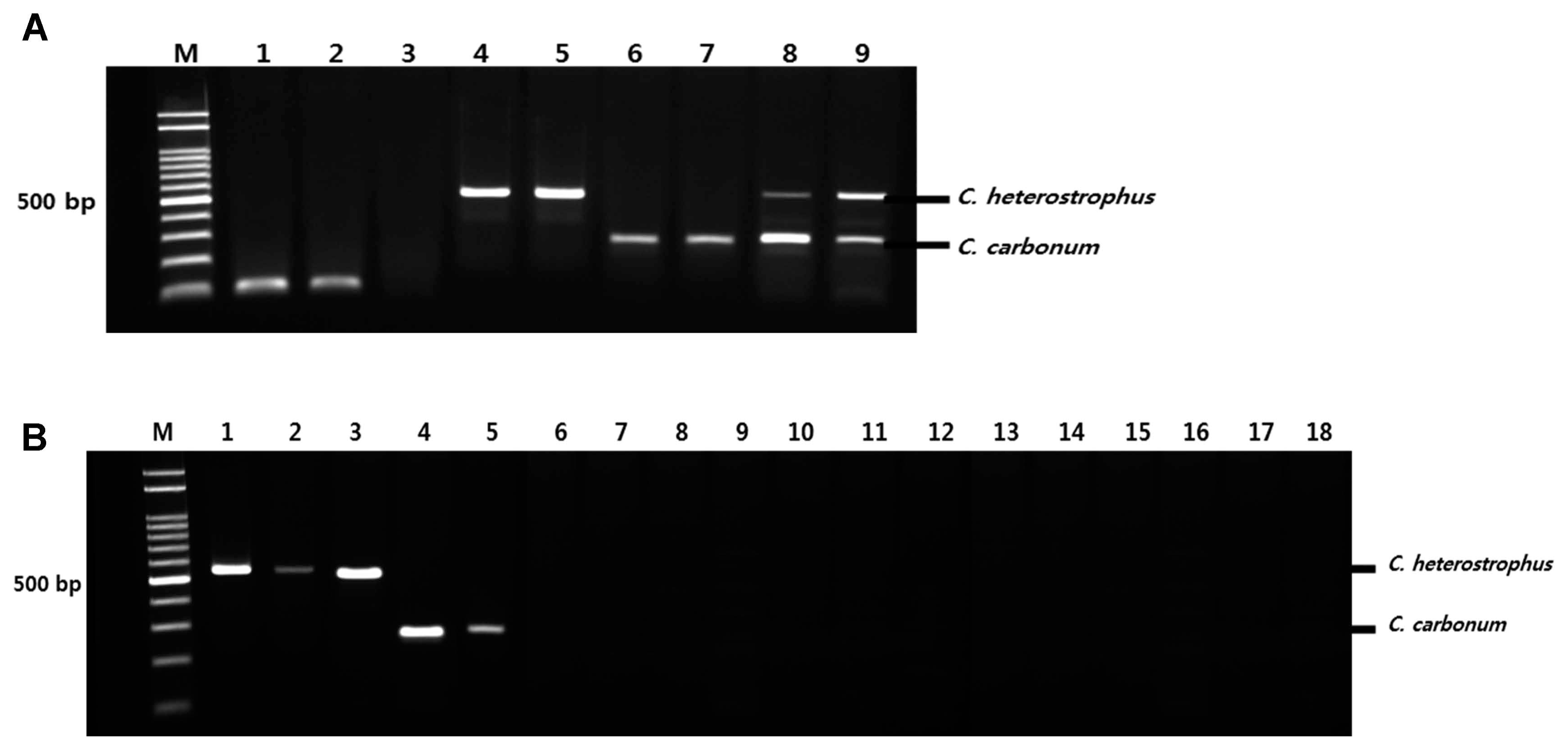
Fig. 4
Sensitivity (detection limit) of duplex PCR in respective fungal genomic DNA quantities (5 ng, 2.5 ng, 1 ng, 0.5 ng, and 0.05 ng/μl). Different quantities of DNA were used for DNA amplification with specific primers and amplicons. The sensitivity of the detection of fungal pathogens was determined using agarose gel electrophoresis. M: molecular size marker (100-2000 bp), Lanes 1-5: The sensitivity level for Cochliobolus heterostrophus F169, Lanes 6-10: The sensitivity level for Cochliobolus carbonum F170.
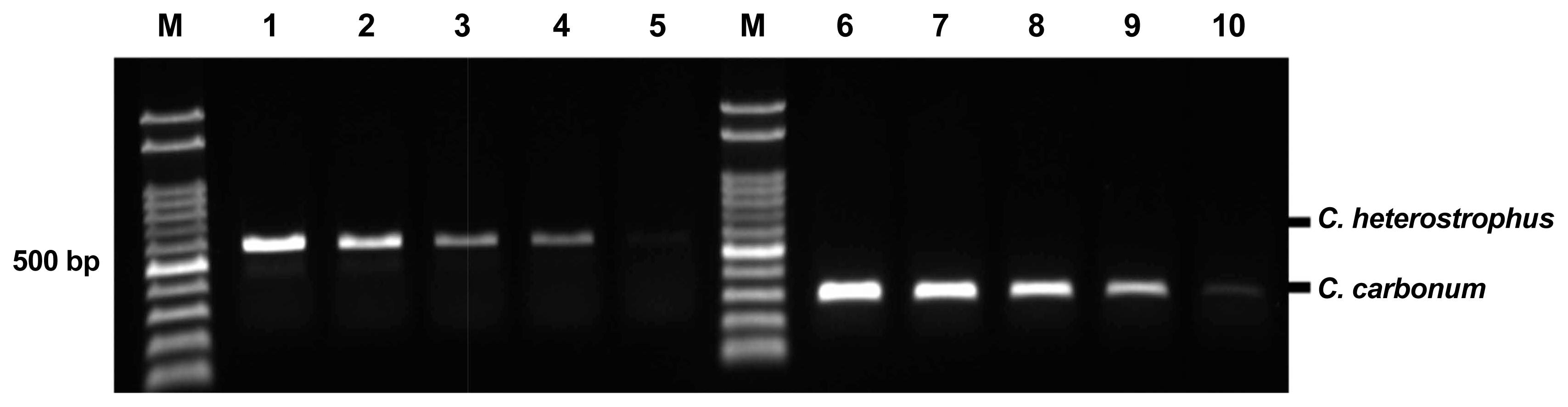
Fig. 5
Symptoms of Cochliobolus heterostrophus F169 and Cochliobolus carbonum F170 artificially inoculated on maize leaves (A) No treatment (left); symptoms of simultaneous inoculation with Cochliobolus heterostrophus F169 and Cochliobolus carbonum F170 (right). (B) Symptoms of inoculation with Cochliobolus heterostrophus F169 (left); symptoms of inoculation with Cochliobolus carbonum F170 (right). Direct detection in maize leaves artificially infected Cochliobolus heterostrophus F169 and Cochliobolus carbonum F170 Lane M: molecular size marker (100-2000 bp), Lane 1: No treatment; Lanes 2-4: Asymptomic corn leaves; Lanes 5-7: Cochliobolus heterostrophus F169-infected; Lanes 8-10: Cochliobolus carbonum F170-infected; Lanes 11-13: Cochliobolus heterostrophus F169 and Cochliobolus carbonum F170-both infected.
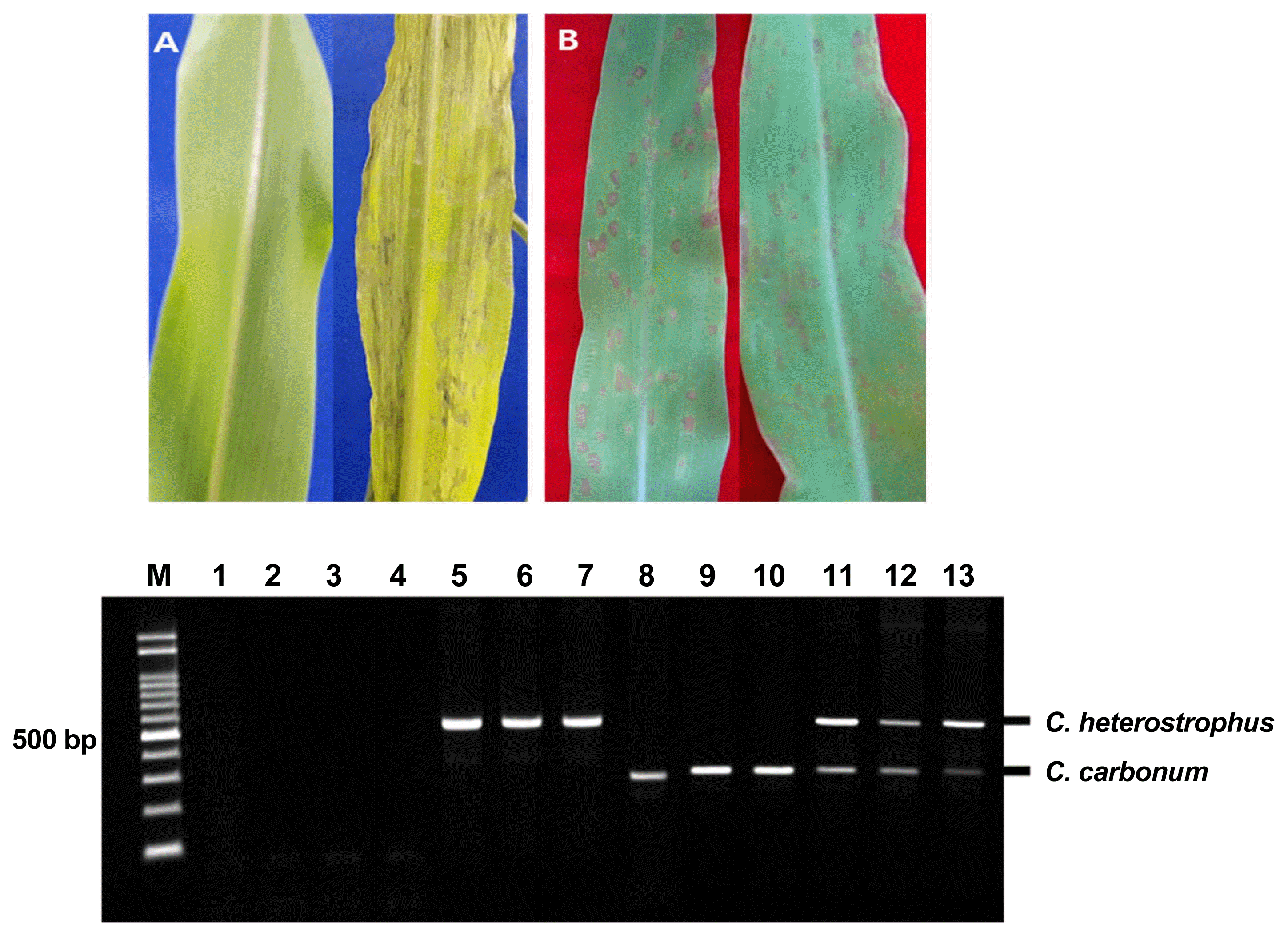
Table 1
Duplex PCR primers to detect two fungal pathogens of corn
| Primers | Primer sequences (5′-3′) | Size (bp) | Source | description |
|---|---|---|---|---|
| Bz-F | GAGAATACCGACCATGTGG | 268 | Cochliobolus carbonum | Northern corn leaf spot |
| Bz-R | TATCTTTAGCTTCCTGTTTGGTC | |||
| Bm-F | TCTCGACAAGCAAATCAAAC | 539 | Cochliobolus heterostrophus | Southern corn leaf blight |
| Bm-R | AGATGATTGCAGTGGTGTTG | |||
| EF1_F | GGCTTTCACCGACTACCCTCCTCT | 91 | Lysøe et al., 2009 | Eukaryotic translation elongation factor 1 alpha |
| EF1_R | ACTTCTCGACGGCCTTGATGACAC |
Table 2
Fungal strains collected from different locations in Korea
References
Ahmadpour, A, Heidarian, Z, Donyadoost-Chelan, M, Javan-Nikkhah, M and Tsukiboshi, T 2012. A new species of Bipolaris from Iran. Mycotaxon. 120:301-307.

Ali, F, Muneer, M, Rahman, H, Noor, M, Shahwar, D, Shaukat, S and Yan, J 2011. Heritability estimates for yield and related traits based on testcross progeny performance of resistant maize inbred lines. J Food Agric Environ. 99:438-443.
Asano, T, Senda, M, Suga, H and Kageyama, K 2010. Development of multiplex PCR to detect five Pythium species related to turfgrass diseases. J Phytopathol. 158:609-615.

Berbee, ML, Pirseyedi, M and Hubbard, S 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 91:964-977.

Da Cunha, KC, Sutton, DA, Fothergill, AW, Cano, J, Gené, J, Madrid, H, De Hoog, S, Crous, PW and Guarro, J 2012. Diversity of Bipolaris species in clinical samples in the United States and their antifungal susceptibility profiles. J Clin Microbiol. 50:4061-4066.



Feng, X, Zhang, X and Yan, D 2012. Detection to pathogens of poplar cankers by the multiplex PCR technique. Scientia Silvae Sinicae. 48:72-77.
Grote, D, Olmos, A, Kofoet, A, Tuset, JJ, Bertolini, E and Cambra, M 2002. Specific and sensitive detection of Phytophthora nicotaianae by simple and nested-PCR. Eur J Plant Pathol. 108:197-207.
Hameed, A, Iqbal, Z, Asad, S and Mansoor, S 2014. Detection of multiple potato viruses in the field suggests synergistic interactions among potato viruses in Pakistan. Plant Pathol J. 30:407-415.



Henson, JM and French, R 1993. The polymerase chain reaction and plant disease diagnosis. Annu Rev Phytopathol. 31:81-109.


Horwitz, BA, Condon, BJ and Turgeon, BG 2013. Cochliobolus heterostrophus: a dothideomycete pathogen of maize. In: Genomics of Soil-and Plant-Associated Fungi, eds. by BA Horwitz, PK Mukherjee, M Mukherjee and CP Kubicek, 213-228. Springer, Berlin, Heidelberg.

James, D, Varga, A, Pallas, V and Candresse, T 2006. Strategies for simultaneous detection of multiple plant viruses. Can J Plant Pathol. 28:16-29.
Kang, JW 2013. Gangwon-do waxy corn seed industrialization plan. Res. Inst. Gangwon, Chuncheon, Korea. 15.(in Korean)..
Kim, YS, Kang, IJ, Shin, DB, Noh, JH, Jeong, JG, Heu, SG and Shim, HK 2017 Corn Cultivation to Reduce the Mycotoxin Contamination. Res Plant Dis. 23:256-261 (in Korean with English abstract).

Korean Society of Plant Pathology. 2009. Corn. In: List of Plant Disease in Korea, eds. by W-G Kim and HM Koo, 53-60 5th ed. Suwon, Korea. (in Korean).
Lysøe, E, Bone, KR and Klemsdal, SS 2009. Real-time quantitative expression studies of the zearalenone biosynthetic gene cluster in Fusarium graminearum. Phytopathology. 99:176-184.


Manamgoda, DS, Cai, L, McKenzie, EH, Crous, PW, Madrid, H, Chukeatirote, E, Shivas, RG, Tan, YP and Hyde, KD 2012. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Diversity. 56:131-144.

Schoch, CL, Seifert, KA, Huhndorf, S, Robert, V, Spouge, JL, Levesque, CA and Chen, W Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 109:6241-6246.


Shin, JH, Han, JH, Lee, JK and Kim, KS 2014. Characterization of the maize stalk rot pathogens Fusarium subglutinans and F. temperatum and the effect of fungicides on their mycelial growth and colony formation. Plant Pathol J. 30:397-406.



Shoemaker, RA 1959. Nomenclature of Drechslera and Bipolaris, grass parasites segregated from Helminthosporium. Can J Bot. 37:879-887.

Shurtleff, MC 1980. Compendium of Corn Diseases. 2nd ed. the Disease Compendia Series. The American Phytopathology Society, Saint Paul, Minnesota, USA. 105.
Sivanesan, A 1987. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. CAB International, Wallingford, UK. 261.
Tan, YP, Crous, PW and Shivas, RG 2016. Eight novel Bipolaris species identified from John L. Alcorn’s collections at the Queensland Plant Pathology Herbarium (BRIP). Mycol Prog. 15:1203-1214.

Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei, M and Kumar, S 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731-2739.



Wang, M, Wang, S, Ma, J, Yu, C, Gao, J and Chen, J 2017. Detection of Cochliobolus heterostrophus races in South China. J Phytopathol. 165:681-691.

Wang, Z, Yang, M, Yang, Y-Q, Peng, Z-K and Zhou, E-X 2010 Morphological and molecular identification of citrus anthracnose pathogen from Guangdong province. Mycosystema. 29:488-493 (in Chinese with English abstract).



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




