A Small GTPase RHO2 Plays an Important Role in Pre-infection Development in the Rice Blast Pathogen Magnaporthe oryzae
Article information
Abstract
The rice blast pathogen Magnaporthe oryzae is a global threat to rice production. Here we characterized RHO2 gene (MGG_02457) that belongs to the Rho GTPase family, using a deletion mutant. This mutant ΔMorho2 exhibited no defects in conidiation and germination but developed only 6% of appressoria in response to a hydrophobic surface when compared to the wild-type progenitor. This result indicates that MoRHO2 plays a role in appressorium development. Furthermore, exogenous cAMP treatment on the mutant led to appressoria that exhibited abnormal morphology on both hydrophobic and hydrophilic surfaces. These outcomes suggested the involvement of MoRHO2 in cAMP-mediated appressorium development. ΔMorho2 mutation also delayed the development of appressorium-like structures (ALS) at hyphal tips on hydrophobic surface, which were also abnormally shaped. These results suggested that MoRHO2 is involved in morphological development of appressoria and ALS from conidia and hyphae, respectively. As expected, ΔMorho2 mutant was defective in plant penetration, but was still able to cause lesions, albeit at a reduced rate on wounded plants. These results implied that MoRHO2 plays a role in M. oryzae virulence as well.
Body
Magnaporthe oryzae, an ascomycete plant pathogenic fungus, causes severe loss of rice production every year that is equivalent to the amount that can feed 60 million people (Han et al., 2015; Kim et al., 2009; Talbot et al., 1993). Utilization of suitable resistant cultivars is considered an effective strategy to control rice blasts. However, M. oryzae overcomes host resistance within several years after their initial appearance in rice paddy fields (Saleh et al., 2014; Zhu et al., 2017). The rice blast caused by M. oryzae starts when germ tubes of conidia emerge on rice leaves (Hamer and Talbot, 1998). Subsequently, a domeshaped specialized infection structure called appressorium develops on the tip of the germ tube before the rice cuticle is penetrated (Dean, 1997; Ryder and Talbot, 2015). The buildup of turgor pressure (up to 8 MPa) inside the appressorium is a key prerequisite for penetration of plant cell walls (Dean et al., 2005). After successful invasion and colonization, asexually reproduced conidia disseminate to uninfected plants. M. oryzae is a polycyclic pathogen and is able to infect almost all tissues of rice plants, which accounts for the severe loss of rice production during favorable conditions (Han et al., 2015; Kim and Lee, 2012).
Regulation of morphogenesis is essential for successful pathogenic development in fungi. The Rho protein family, which was first identified in Aplysia species, belongs to the family of small GTPases (Madaule and Axel, 1985; Ridley, 2001). Most Rho proteins are GTP-hydrolyzing molecular switches, which participate in different signaling pathways for cell growth, morphogenesis, and gene regulation (Arellano et al., 1999; Etienne-Manneville and Hall, 2002). Rho GTPases contain five highly conserved motifs (G1–G5) and two functional elements, switch I between G1 and G2 and switch II between G2 and G3 (Smithers and Overduin, 2016). The G1–G5 loops form nucleotide-binding sites, regulating GTP hydrolysis (Hanna and El-Sibai, 2013). The switch I element is a core effector that is important for the interaction with downstream partners (Karnoub et al., 2004). The switch II element coordinates nucleophilic water that is required for GTPase activity (Schaefer et al., 2014). A special helical amino acid sequence of approximately 10–15 residues located between G4 and G5 distinguishes the Rho protein family from other GTPases. This helical sequence has no effect on the function of GDP or GTP binding but modulates Rho protein activity through an interaction with guanine nucleotide exchange factors, thus affecting downstream effectors (Wittinghofer and Vetter, 2011).
In the fungal kingdom, Rho proteins modulate diverse biological processes such as growth, differentiation, secretion, vesicular trafficking, and transcription. Several members of the Rho family have been characterized in fungal pathogens, including RHO1, RHO2, RHO3, RHO4, CDC42, and RAC1 (Dean et al., 2005; Du et al., 2013). RHO1 gene helps control cell wall synthesis and aggregation of actin cables in Fusarium oxysporum, Aspergillus nidulans, and Saccharomyces cerevisiae (Guest et al., 2004; Martínez-Rocha et al., 2008; Ridley, 2001). RHO2 is involved in regulating the cell wall integrated pathway in Neurospora crassa and Colletotrichum gloeosporioides (Richthammer et al., 2012; Xu et al., 2016). RHO3 is associated with the regulation of mitochondria distribution, conidial morphology, germination, and vegetative growth in M. oryzae, Trichoderma reesei, and Botrytis cinerea (An et al., 2015; Vasara et al., 2001; Zheng et al., 2007). RHO4 is involved in septa formation by regulating the actin cytoskeleton in fission yeast and N. crassa (Nakano et al., 2003; Rasmussen and Glass, 2005). In Ustilago maydis, CDC42 serves as an effector of cell separation during budding, and RAC1 is necessary for the switch from budding to hyphal growth (Mahlert et al., 2006). However, the detailed functions of RHO2 in M. oryzae have not been identified.
In this study, we functionally characterized MoRHO2 using a targeted gene deletion in M. oryzae. Our results indicate that deletion of MoRHO2 does not affect vegetative growth, conidiation, or conidial germination, but it leads to a delayed development of appressoria on hydrophobic surfaces. Exogenous cAMP treatment only partially rescues morphologically abnormal development of appressoria. Taken together, our data suggest that MoRHO2 plays an important role in pre-infection development in M. oryzae.
Materials and Methods
Fungal strains and culture conditions
The wild type (KJ201) and transformants obtained in this study were grown on medium that included oatmeal (50 g/l oatmeal and 25 g/l agar powder), complete medium (6 g/l yeast extract, 6 g/l casamino acids, 10 g/l sucrose, and 15 g/l agar powder) and V8 medium (80 ml V8 juice, 310 μl 10 N NaOH solution, and 15 g/l agar powder) at 25°C with constant light. The conidial suspension was prepared by filtrating conidia obtained from 7-day-old V8 medium through two layers of Miracloth (Calbiochem, San Diego, CA, USA). Mycelia grown in liquid conditioned medium (CM) for 8 days were used to extract DNA and RNA.
Sequence and phylogenetic analysis
We obtained all sequences from online databases of the Broad Institute of MIT and Harvard (http://www.Broadinstitute.org), the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and the Comparative Fungal Genomics Platform (http://cfgp.riceblast.snu.ac.kr) (Park et al., 2007). MEGA 6.0 was used to analyze phylogenetic relationship of MoRHO2 with orthologs of other organisms. Domain structures of the Rho protein family were analyzed with InterPro Scan, version 53 (http://www.ebi.ac.uk/interpro/), and visualized using Illustrator for Biological Sequences, version 1.0.3. All primers in this study were designed by Primer Quest Design Tool (http://sg.idtdan.com/site).
RNA isolation and gene expression analysis
The RNA (5 μg) used to synthesize cDNA, was extracted from mycelia, conidia, germinated conidia, appressoria and invasive hyphae in rice leaves, by using Easy-SpinTM. RT-PCR was conducted to detect transcripts of MoRHO2 in transformants with RTF/RTR primers (Han et al., 2015). The process was performed in a 20-μl mixture containing 1 μl cDNA (25 μg/μl), 1 μl forward primer, 1 μl reverse primer, 4 μl Pfu polymerase, and 13 μl sterilized distilled water. In total, 30 cycles of RT-PCR were run in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA). qRT-PCR was performed to compare the expression levels of MoRHO2 during development of M. oryzae as previously described (Han et al., 2015). The reaction was performed using a 10-μl mixture containing 1 μl cDNA, 1 μl forward primer, 1 μl reverse primer, and HIPI Real-Time PCR 2×Master Mix (SYBR Green; Molecular Probes, Eugene, OR, USA). PCR was performed with qRTF and qRTR primers using the One Step Real-Time PCR System (Applied Biosystems). Cycle threshold (Ct) was normalized to that of the β-tubulin gene (MGG00604) and treated samples as 2−ΔCt, where −ΔCt = (Ct, target gene – Ct, β-tubulin). Fold changes in expression were calculated as 2−ΔΔCt, where −ΔΔCt = (Ct, target gene – Ct, β-tubulin)test condition – (Ct, WT – Ct, β-tubulin)CM. Three replicates were conducted to confirm the results of qRT-PCR.
Targeted deletion of MoRHO2 and construction of complementation strains
MoRHO2 targeted deletion construct was generated by double-joint PCR (Choi et al., 2011; Yu et al., 2004). Upstream and downstream flanking regions (1.5 kb) of the target gene were amplified by primers 3F/3R and 5F/5R (Table 1). The HPH cassette was amplified using primers HPH-F/HPH-R. The three PCR products were fused with primers 5F/3R. The final deletion construct was amplified by primers NF/NR, and subsequently transformed into wild-type protoplasts (Kim et al., 2009). Hygromycin-resistant transformants were selected on TB3 medium containing Hygromycin B (200 μL/ml). Southern blotting and RT-PCR were used to confirm the targeted deletions. For complementation, the targeted gene was amplified from the wild-type genome by primers cmF/cmR. The amplicon was used to co-transform protoplasts of the deletion mutant with the pII99 vector containing a Geneticin-resistance cassette.
Appressorium formation on the hydrophobic surface
Conidia grown on 6-day-old V8 medium were harvested by adding 5 mL sterilized, distilled water and filtering through two layers of Miracloth (Calbiochem). The conidial suspension was centrifuged at 5,000 rpm for 10 min, followed by removal of the supernatant, and was diluted to a final concentration of 5 × 104 conidia/ml (Han et al., 2015; Kim et al., 2014). Drops of conidial suspension (20 μl) were placed on hydrophobic coverslips and hydrophilic slide glasses with or without the addition of exogenous 5 mM cAMP and incubated in moist boxes. Appressoria formed on germinated conidia were calculated by counting at least 100 conidia per replicate in at least three independent experiments with three replicates per experiment.
Wound inoculation and sheath assays
Three-week-old seedlings of the susceptible rice cultivar Oryza sativa cv. Nakdongbyeo were used to conduct leaf infection assays (Huh et al., 2017). Conidial suspensions (5 × 104 conidia/ml) of wild-type, mutant, and complemented strains collected from 10-day-old oatmeal medium were sprayed onto rice leaves and incubated in the dark for 24 h, then moved to a growth chamber for a 16 h photoperiod with fluorescent light (Kim et al., 2014). Disease development was examined after 7 days of incubation. Appressorium penetration and hyphal infection assays were performed using dropping conidial suspensions (5 × 104 conidia/ml) or by placing agar plugs (6 mm in diameter) of wildtype, mutant, and complemented strains onto rice leaves incubated under the same conditions as the spray assay. Infected tissues of rice leaves were observed after 2 days of inoculation. Briefly, a conidial suspension (5 × 104 conidia/ml) was dropped on a rice sheath and incubated, then the invasive growth in plants was observed after 2 days.
Results
Phylogenetic analysis and domain structure of the Rho family
The M. oryzae genome contains six members of the Rho family, which include MoRHO1 (MGG_07176), MoRHO2 (MGG_02457), MoRHO3 (MGG_10327), MoRHO4 (MGG_03901), MoCDC42 (MGG_00466), and MoRAC1 (MGG_02731). Each of the six protein sequences consists of one conserved domain named the small GTP-binding protein domain (IPR005225) (Fig. 1A). A BLASTP search of the NCBI database showed that MoRHO2 had 64% identity with MoRHO1, 47% identity with MoRHO3 and MoCDC42, 49% identity with MoRHO4, and 53% identity with MoRAC1 (Fig. 1B). A phylogenetic tree was constructed to analyze the relationships among MoRHO2 and orthologs from other organisms (Fig. 2A). MoRHO2 was closely related to proteins from C. gloeosporioides, F. graminearum, N. crassa, S. sclerotiorum, and A. nidulans, but was genetically distant to other homologs from animal or plants. The small GTPase MoRHO2 shared 92% sequence identity with CEF78997.1 from F. graminearum and 27% sequence identity with related proteins from D. melanogaster (Fig. 2B). Together, these results indicate that RHO2 orthologs are well-conserved in fungi.

Domain structure and conserved amino acids of RHO GTPase in M. oryzae. (A) Schematic structure of the small GTP-binding protein domain (IPR005225) in the RHO protein family. The domain structure was predicted using InterProScan. (B) The conserved amino acid sequence alignment of Rho GTPase. G1, G2, G3, G4, G5, switch I, and switch II denote special motifs in the small GTPbinding protein domain. The identity of each protein BLAST search with MoRHO2 is followed by its name.
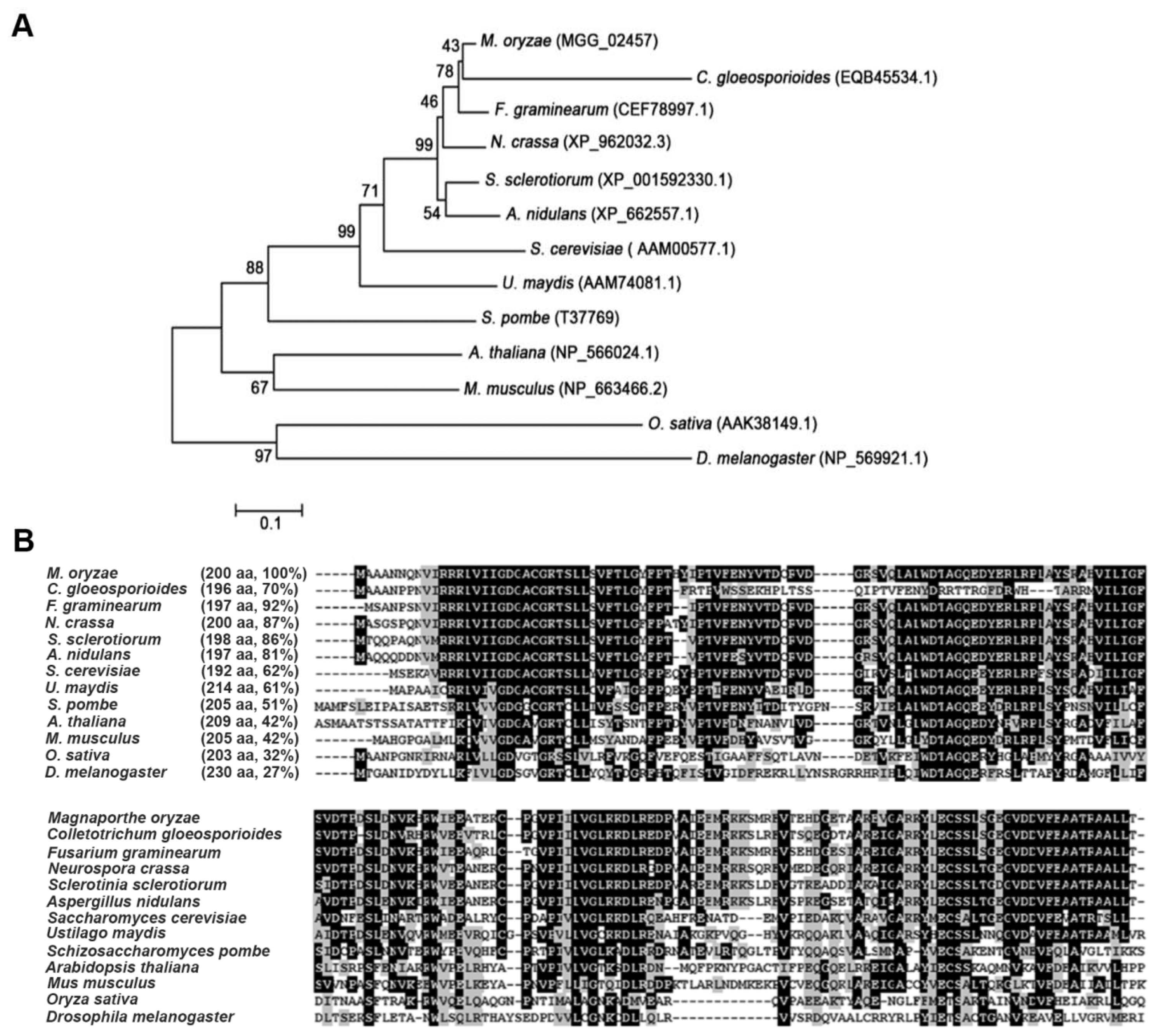
Phylogenetic analysis and conserved amino acid sequence alignment of MoRHO2 and homologues from other organisms. (A) Phylogenetic analysis among MoRHO2 and homologues. A phylogenetic tree was generated using a neighbor-joining method based on comparing MoRHO2 and its homologues. (B) The conserved amino acid sequence alignment among MoRHO2 and homologues. The identity of each protein BLAST search with MoRHO2 is followed by its name.
Expression and targeted gene deletion of MoRHO2.
To order to predict functional roles of MoRHO2 in developmental stages of M. oryzae, the expressions of targeted genes were measured using qRT-PCR during the interaction of M. oryzae and its host plant. MoRHO2 was significantly expressed in conidia, appressoria, and invasive hyphae in host plants, but not in germinated conidia (Fig. 3A). Expression of MoRHO2 was greatly induced (> 8-fold) in infected host plants, raising the possibility that MoRHO2 may play a role in pre- and post-infection. Therefore, we generated a targeted gene deletion mutant to study the roles of MoRHO2 in fungal pathogenic development. The deletion mutant was generated by introducing a knockout construct selected through the selectable marker-hygromycin cassette (Fig. 3B). In addition, ΔMorho2 was further verified by Southern blotting and RT-PCR (Fig. 3C and D). To confirm that the phenotype of ΔMorho2 resulted from deletion of the targeted gene, the mutant was complemented with a genomic copy of MoRHO2. A complemented transformant (Morho2c) showed recovered expression of MoRHO2.
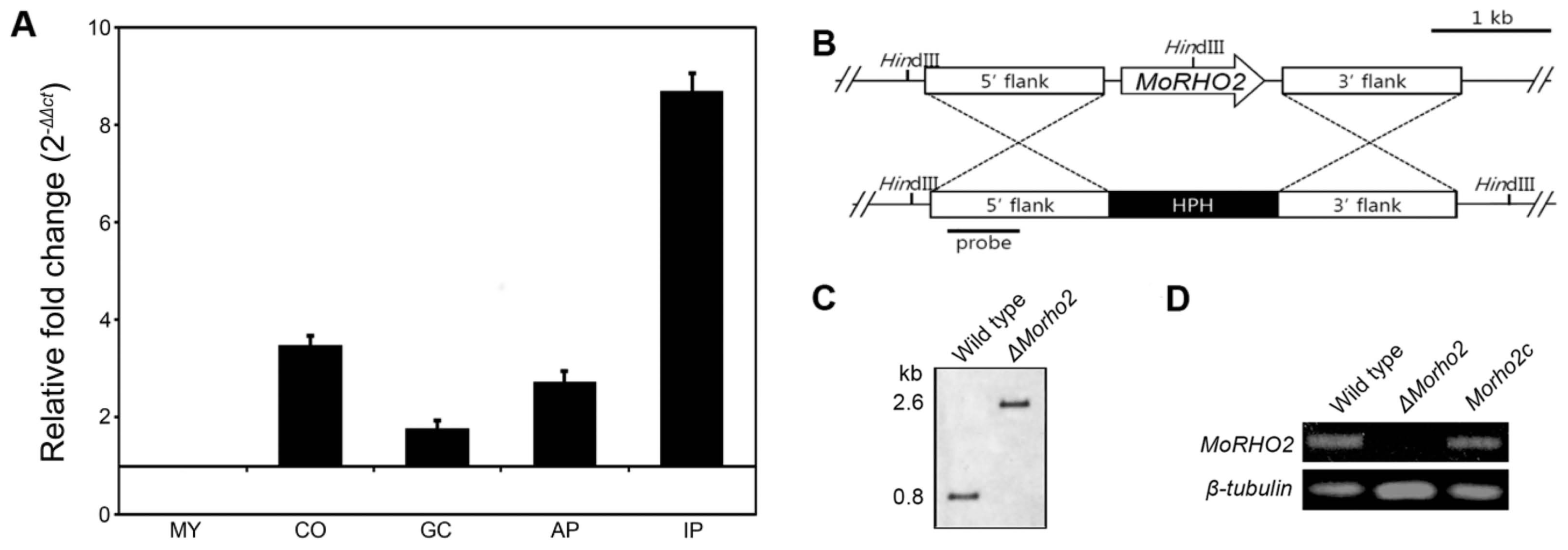
The expression profile and targeted gene deletion of MoRHO2. (A) The expression profile of MoRHO2 in different developmental stages of M. oryzae. The expression of MoRHO2 was measured during five stages including mycelia (MY), conidia (CO), germinated conidia (GC), appressoria (AP), and infectious hyphae stages in rice leaves (IP). The results were normalized to β-tubulin and presented with a relative value of 1 in MY. (B) The targeted gene knockout of MoRHO2. The knockout strategy used the HPH cassette to replace MoRHO2. (C) The conformation of the MoRHO2 deletion using southern blot analysis. The genomic DNA was digested with HindIII and hybridized with specific probes. (D) Reverse transcription-PCR was used to check the expression of MoRHO2. The total RNA was extracted from wild type, ΔMorho2, and Morho2c samples.
Appressorium formation on a hydrophobic surface
We evaluated the role of MoRHO2 gene in vegetative growth rate, conidiation, and conidium germination in M. oryzae by comparing phenotypes of the wild type, ΔMorho2, and Morho2c strains. The ΔMorho2 mutant exhibited no significant differences compared to the wildtype and Morho2c in vegetative growth, conidiation, and conidium germination, which indicated that MoRHO2 gene is not associated with these developmental stages (data not shown). Then we tested the role of MoRHO2 in appressorium development. When the conidial suspension was dropped onto hydrophobic coverslips, 92.6% and 99% of the wild type conidia generated appressoria at 8 and 48 hr, respectively (Fig. 4). Surprisingly, ΔMorho2 did not develop appressorium on hydrophobic surface at 8 hr, and only 6% at 48 hr. This result suggested that MoRHO2 is required for proper development of appressoria in response to the hydrophobic surface. Because C. gloeosporioides RhoB, a MoRHO2 orthologous, is involved in the cAMP signaling pathway (Xu et al., 2016), we further investigated the effects of exogenous cAMP on defective appressoria development in ΔMorho2. Notably, in the presence of cAMP, appressorium formation was restored in ΔMorho2 like wild type on hydrophobic surface. However, these appressoria were abnormally shaped, indicating that MoRHO2 is important for surface recognition and morphological regulation during appressorium development.

Appressorium formation on artificial surfaces. (A) Statistical analysis of appressoria formed on the hydrophobic and hydrophilic surface. Appressorium formation was assessed at 48 h after inoculation. (B) The appressorium morphology on a hydrophobic surface. Appressoria were observed after a 6 h incubation. Scale bar = 20 μm.
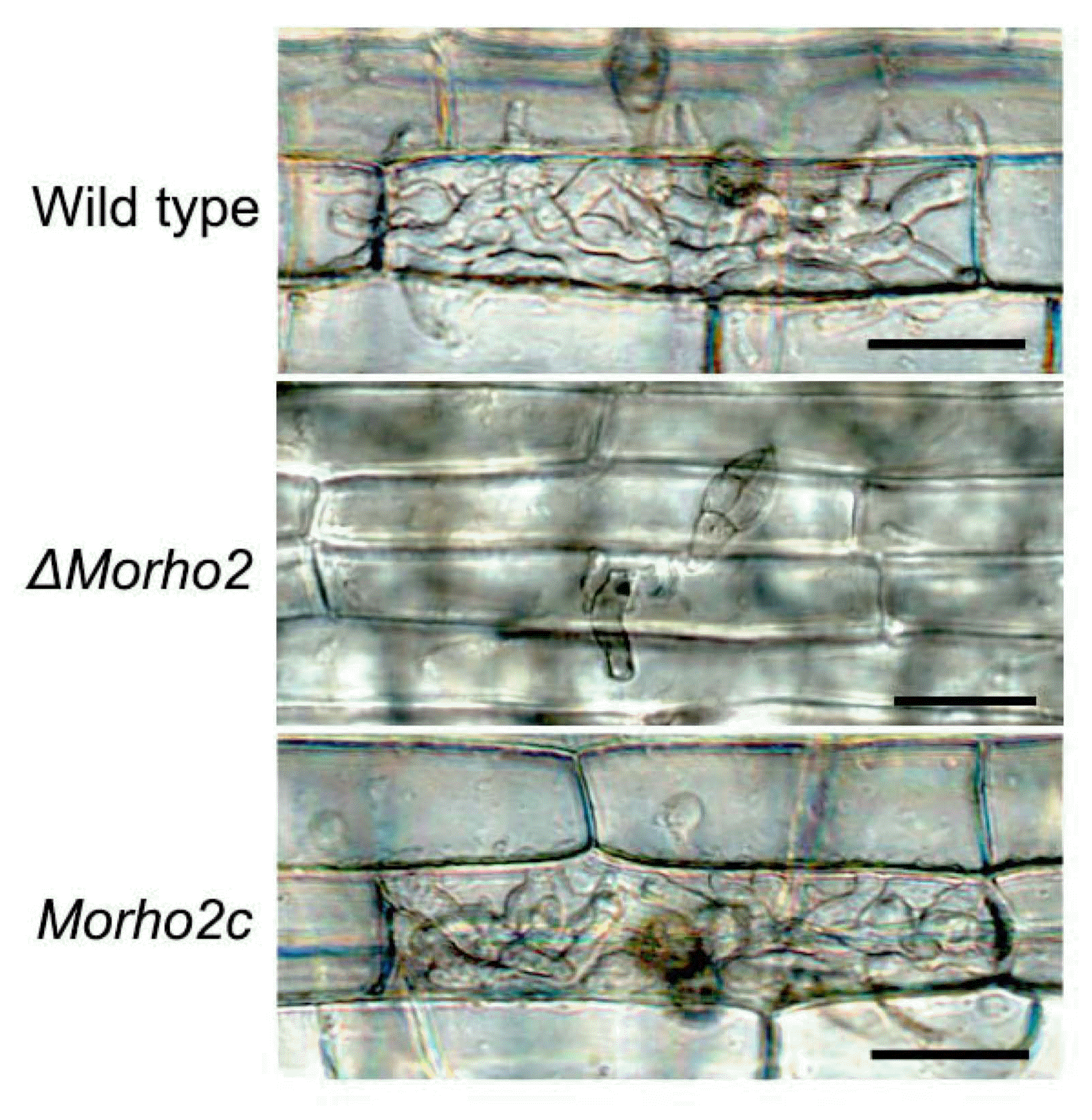
In a previous study, M. oryzae penetrated host cells through appressorium-like structure (ALS) developed on hyphal tips (Kong et al., 2013). Therefore, we examined whether MoRHO2 was involved in ALS development in M. oryzae. The ΔMorho2 mutant showed a delayed development of ALS with an abnormal shape from hyphal tips on the same hydrophobic surface. These results suggested that MoRHO2 is required for appressorium morphogenesis derived from conidia and hyphae.
Roles of MoRHO2 in pathogenic development
To understand the role of the MoRHO2 gene in disease development, susceptible 3-week-old rice leaves were sprayed with conidial suspensions of wild type, ΔMorho2, and Morho2c. Treatment of wild type and Morho2c led to typical necrotic lesions (Fig. 6A). In contrast, ΔMorho2 completely failed to cause disease symptoms. To further characterize the roles of MoRHO2 in penetration, conidial drops were inoculated onto detached wounded or unwounded rice leaves (Fig. 6B). Similar to the results from conidial spray assays, wild-type and Morho2c strains developed typical lesions on unwounded rice leaves, but ΔMorho2 did not cause any lesions. However, ΔMorho2 led to lesion development on wounded rice leaves, although they were very weak compared to those of wild type or Morho2c. When hyphal plugs were inoculated onto detached rice leaves, ΔMorho2 caused weaker disease symptoms on both wounded and unwounded rice leaves compared to those by the wild type and Morho2c. Together, these results suggested that MoRHO2 is required for full virulence in M. oryzae.
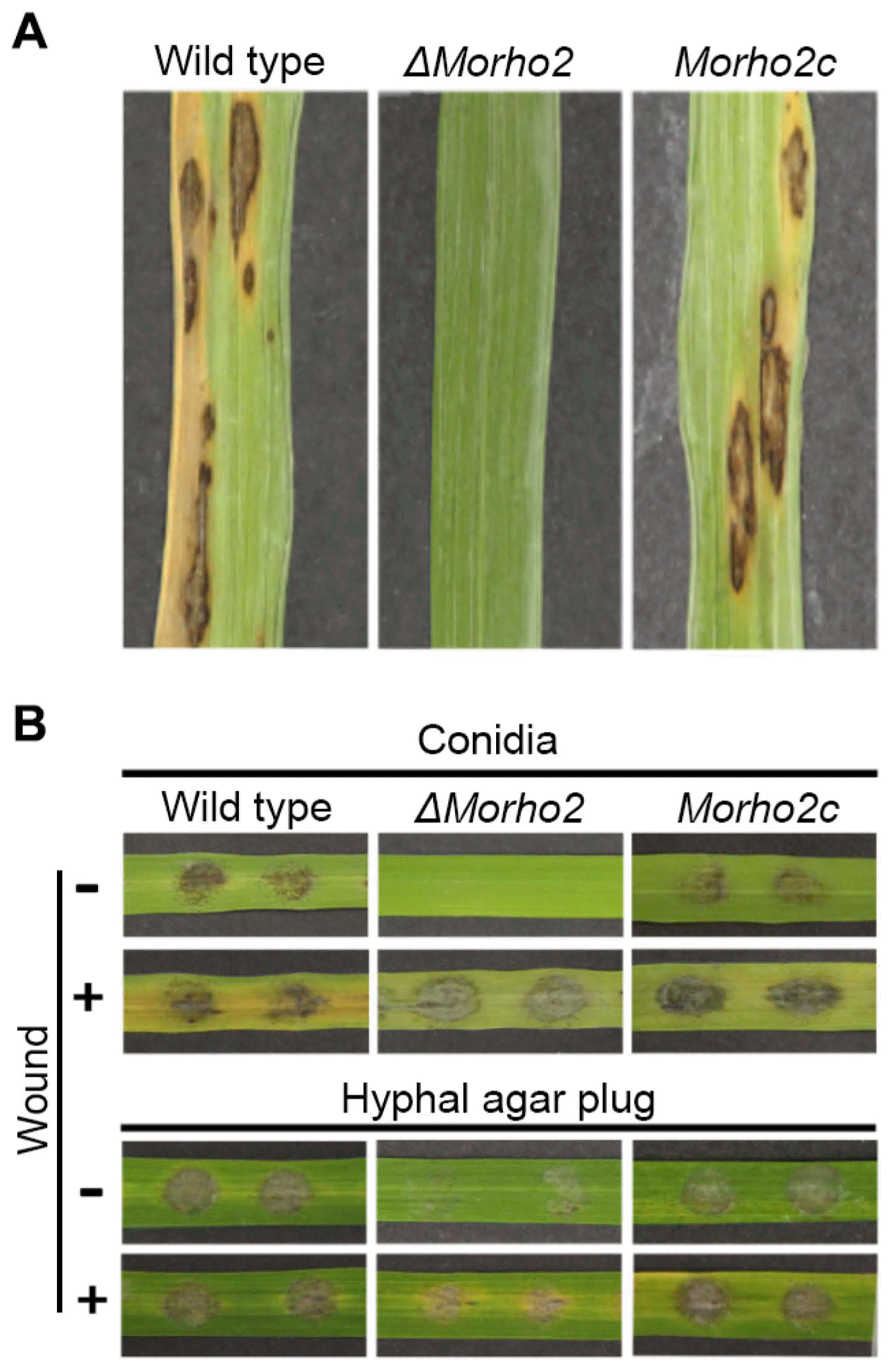
Plant pathogenic assays. (A) The spray assays. The conidial suspension was sprayed onto rice leaves and the leaves were incubated for 7 days. (B) The influence of wounding on disease development. Conidial drops or hyphal plugs (6 mm in diameter) were inoculated onto rice leaves with or without wounding and the leaves were incubated for 2 days.
Appressorium penetration and invasive hyphal growth
To further characterize the association between MoRHO2 and disease development, we tested ΔMorho2 for penetration and invasive growth by inoculating a conidial suspension onto rice sheath cells. Similar to the defective appressorium development on artificial surfaces, conidia of ΔMorho2 were unable to develop appressoria on rice sheath cells, in contrast to those of the wild type and Morho2c. A small number of conidia (< 1%) in ΔMorho2 developed appressoria but these were morphologically abnormal and were unable to penetrate rice cells (Fig. 7). These results strongly suggest that MoRHO2 is required for appressorium-mediated penetration in M. oryzae.
Discussion
M. oryzae is a model organism for studying fungal development and pathogenic development (Valent, 1990). In the present study, we showed that MoRHO2 plays an important role in M. oryzae preinfection-related development. MoRHO2 encodes a GTP-binding protein, which belongs to the Rho GTPase family (Dean et al., 2005). To characterize the functional role of MoRHO2 in M. oryzae, we generated a MoRHO2 deletion mutant and compared phenotypes during developmental stages in M. oryzae. The ΔMorho2 mutant was unaffected in vegetative growth, conidiation, and conidial germination compared to the wild type, which indicates that MoRHO2 is not associated with these developmental stages.
The vast majority of ΔMorho2 conidia failed to produce appressoria on hydrophobic surface, indicating that MoRHO2 is required for appressorium formation in response to hydrophobicity (Fig. 4). Because the cAMP signaling pathway is well known in appressorium formation, we tested the effect of exogenous cAMP in the appressorium formation (Xu and Hamer, 1996). The appressorium formation rate in ΔMorho2 increased to a level similar to that wild type, but all of the ΔMorho2 appressoria exhibited an abnormal morphology, which suggests that MoRHO2 is involved in the recognition of hydrophobicity for proper development of appressoria. However, the treatment of exogenous cAMP enabled ΔMorho2 mutant to bypass the signal recognition for appressorium development, but not for appressorium morphogenesis. It is noteworthy that the deletion of CgRhoB in Colletotrichum gloeosporioides, a MoRHO2 ortholog, leads to the abnormal increase in cAMP levels, enhancement in appressorium development and alteration in cell wall integrity (Xu et al., 2016). Therefore, it is reasonable to hypothesize that MoRHO2 is involved in regulating cAMP level for appressorium development in M. oryzae, as supported with restoration of appressorium development with exogenous cAMP treatment in ΔMorho2 (Fig. 4). However, the mechanistic function of MoRHO2 in the cAMP signaling pathway is still unknown. M. oryzae develops an appressorium-like structure (ALS) on hyphal tips (Kong et al., 2013). Similar to abnormal appressoria on conidial germ tubes, the ΔMorho2 mutant also developed morphologically abnormal ALS on hyphal tips (Fig. 5). These data indicate that MoRHO2 is essential for appressorium morphogenesis on tips of conidial germ tubes and hyphae tips. Interestingly, the ΔCgRhoB mutant exhibited shorter germ tubes, reduction in conidial germination while the ΔMorho2 mutant was normal in germ tube formation and conidial germination, suggesting divergent roles of RHO2 orthologs in fungal pathogens (Xu et al., 2016).

An appressorium-like structure (ALS) formed on hyphal tips. Hyphal plugs (5 mm in diameter) of wild type, ΔMorho2, and Morho2c samples were placed on hydrophobic surfaces. Photographs were taken after 24 and 36 h. Scale bar = 50 μm.
Abnormal appressoria morphology in response to exogenous cAMP in the ΔMorho2 mutant may suggest a defect in turgor pressure. This is supported by the lack of symptom development in both spray assay and sheath assay (Fig. 6). Interestingly, other studies have reported roles of MoRHO2 orthologs in the cell integrity in A. niger, F. graminearum, and N. crassa (Kwon et al., 2011; Richthammer et al., 2012; Zhang et al., 2013). The importance of cell wall integrity in maintaining turgor pressure of appressoria in M. oryzae is well documented (Xu et al., 1998). Moreover, in the budding yeast Saccharomyces cerevisiae, Rho2 affects Slt2, a mitogen-activated protein kinase (MAPK), through protein kinase C, thus controlling cell integrity including ionic homeostasis and cell wall maintenance (Matia-González and Rodríguez-Gabriel, 2011). Considering that Mps1, a Slt2 ortholog, is associated with cell wall integrity in M. oryzae, we speculate that RHO2 may affect the regulation of Mps1 for cell wall integrity (Xu et al., 1998).
In summary, we characterized the role of MoRHO2 gene in M. oryzae morphogenesis and disease development. Rho GTPases are involved in essential molecular signaling pathways regulating pathogenicity in fungal species. MoRHO2 gene was involved in preinfection-related development of M. oryzae, including appressorium development and morphogenesis, and invasive growth inside host plant cells. Overall, our results provide a functional basis for MoRHO2-mediated pre-infection development in M. oryzae.
Acknowledgement
This study was supported by Basic Science Research Program through the National Research Foundation of Korea grant (NRF-2013R1A1A1008444) funded by the Ministry of Education, Science and Technology, and by a grant (117035-03-2-SB010) funded by Export Promotion Technology Development Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.