 |
 |
| Plant Pathol J > Volume 35(3); 2019 > Article |
Abstract
In this research, the relationships among the 31 microsatellite markers with charcoal rot disease resistance related indices in 130 different soybean cultivars and lines were evaluated using association analysis based on the general linear model (GLM) and the mixed linear model (MLM) by the Structure and Tassel software. The results of microsatellite markers showed that the genetic structure of the studied population has three subpopulations (K=3) which the results of bar plat also confirmed it. In association analysis based on GLM and MLM models, 31 and 35 loci showed significant relationships with the evaluated traits, respectively, and confirmed considerable variation of the studied traits. The identified markers related to some of the studied traits were the same which can probably be due to pleiotropic effects or tight linkage among the genomic regions controlling these traits. Some of these relationships were including, the relationship between Sat_252 marker with amount of charcoal rot disease, Satt359, Satt190 and Sat_169 markers with number of microsclerota in stem, amount of charcoal rot disease and severity of charcoal rot disease, Sat_416 marker with number of microsclerota in stem and amount of charcoal rot disease and the Satt460 marker with number of microsclerota in stem and severity of charcoal rot disease. The results of this research and the linked microsatellite markers with the charcoal rot disease-related characteristics can be used to identify the suitable parents and to improve the soybean population in future breeding programs.
Soybean (Glycin max L.) is a diploid (2n = 2├Ś = 40), annual and dicotyledon plant from the legumes family and Papilionoideae subfamily which is planted and exploited in both grain and fodder forms (Singh, 2006). Soybean has a special status among the plants due to its high level of protein and oil. Soybean has in average 40% protein and 20% oil which is an unrivaled grain in terms of nutritional value from among the important crops (Bilyeu et al., 2010). In natural and agricultural ecosystems, plants are exposed to the stresses. Various factors including biotic (insects, fungus, viruses and weeds) and abiotic factors (drought, salinity, high and low temperature, flooding and radiation) affect growth of the crop plants. Soybean is sensitive to a large number of pathogens that cause the most damage to seedling and root of plant. One of the soil-borne pathogens which attack the root and crown is the fungus Macrophomina phaseolina (Tassi) Goid which is the factor of charcoal rot pathogen or wilting. This fungus is poly-phage and pollutes more than 500 plant species in 100 families, including the monocotyledon and dicotyledon (Jana et al., 2003). In favourable conditions, this pathogens causes the burning and death of seedling, the crown rot and charcoal rot in most important crops such as soybean, cotton, corn, sunflower, sorghum and so on (Babu et al., 2007). It seems that the only practical way to control the soybean charcoal rot is to use the resistant cultivars (Mengistu et al., 2001). Therefore, identifying the resistant cultivars and mapping resistance gene helps significantly the plant breeders to progress the breeding programs for producing the disease resistant cultivars.
Association analysis which is also known as linkage disequilibrium (LD) mapping, has main advantages that mapping the genes. Since natural populations are used in association analysis, firstly, there is wider genetic diversity than the two-parents populations. Secondly, upon the population, LD mapping has high accuracy, because all meiosis events accumulated during the plant evolutionary history are taken into account, while in the ordinary method, mapping only occur in a number of cross or autogamous generations (Musial et al., 2008). Linkage disequilibrium has a important application in association mapping studies. In case of any linkage disequilibrium between the molecular marker and genes controlling a special trait, significant relationship can be determined between the marker and traits and using it in breeding programs. Therefore, association mapping is of high importance in determining the amount of linkage disequilibrium between the marker and trait (Oraguzie et al., 2007).
Using the association mapping, Iqbal et al. (2001) identified six QTLs for resistance against the sudden death syndrome (SDS) disease in soybean based on the ANOVA method. By using the SSR markers, Wang et al. (2008) identified twelve QTLs related to resistance against the powdery mildew in soybean by association analysis. Li et al. (2010) and Wu et al. (2011) identified 45 QTLs related to the resistance to Phytophthora distributed on 15 chromosomes by using the association analysis. Fusari et al. (2012) and Iquira et al. (2015), identified the QTLs for resistance to Sclerotinia stem rot disease in sunflower and soybean respectively by using the mixed linear model, they identified a candidate gene which justified 20% of phenotypic variance. Through association analysis, Sun et al. (2014) identified four alleles of Satt 634-133, Satt 634-149, Satt 222-168 and Satt 301-190 related to the partial resistance to Phytophthora in soybean. Sonah et al., (2015) between one and eight genomic loci associated with seed weight were identified. Coser et al. (2017), new sources for charcoal rot disease resistance were identified from both field and greenhouse screening from maturity groups I, II, and III. Five significant single nucleotide polymorphism (SNP) and putative candidate genes related to abiotic and biotic stress responses were reported from the field screening; while greenhouse screening revealed eight loci associated with eight candidate gene families, all associated with functions controlling plant defense response.
It seems that genes controlling some morphological attributes and a few number of diseases in soybean have been identified based on the consistency analysis as well as association analysis but genes or QTLs controlling the resistance to the charcoal rot disease in soybean have not been yet mapped. In this study, SSR markers linked to some important traits related to the charcoal rot disease were identified by association analysis in Iranian soybean germplasm.
To evaluate the resistance of different soybean genotypes to charcoal rot, the seeds of 130 soybean genotypes from different maturity groups were planted in two separate experiments as a randomized complete block design with three replications at the research field of Seed and Plant Improvement Research Institute (SPIRI), Karaj Iran, during two years, 2014 and 2015. The plant materials of this study were part of the Iranian soybean germplasm achived from SPIRI (Table 1). The seeds of each genotype were planted in four lines of 2.5 m with a distance of 50 cm between lines and 10 cm between plants. Primary plowing and disk were carried out at a depth of 30 cm and 15 cm, respectively, and the ground levelling was done by the trowel. Nitrogen fertilizer with 150 kg/ha criterion according to the soil test was added to the plots in equal proportions in three stages, before planting, flowering and podding stage. The first irrigation was carried out 3 days before planting and the next irrigations were done once a week. Weed control was carried out manually on several occasions.
The genotypes were inoculated with the pathogen at flowering stage employing Tesso and Ejeta (2011) method with some modifications. For contamination in field conditions, isolate S8, isolated and purified from infected soybean plants at SPIRI field, was propagated on a potato dextrose agar (PDA) culture medium to obtain a three-day culture. Seven mm discs made from the fungus colony margin were placed in the center of 9 cm petri dishes containing a new PDA culture medium. Then, in sterile conditions, four sterilized toothpicks were placed in each petri with the same intervals and on two sides of the mycelium disk. The petri dishes were stored in dark conditions at 30┬░C for 7 days. After the toothpicks were covered with mycelium colonies and fungal microsclerotia, they were transferred to the field for inoculation of plants at the flowering stage. To inoculate, some holes were firstly created on the stems horizontally by an awl which were in the diameter of the toothpick and the contaminated toothpicks were secondly inserted into the plant stem. To determine the resistance and susceptibility of soybean genotypes to charcoal rot disease, the traits including pod weight, grain weight, 100 grain weight, grain yield, number of microsclerota in stem, amount of charcoal rot disease (I) and severity of charcoal rot disease (S) were measured. Amount of charcoal rot disease (I) and severity of charcoal rot disease (S) were calculated based on Eqs. (1) and (2), respectively.
Where n is the number of plants with symptoms of the disease and N is the total number of evaluated plants (Cardoso et al., 2004).
The severity of disease (S) is based on the rate of colour changes in the plant tissue (stem).
Which Hd is the height of the stem discoloration or the length of the lesion, and Ht is the total height of the stem measured (Mengistu et al., 2007). The ruler was used to measure the length of the lesion caused by the fungus.
For genotypic evaluations, 3 to 4 newly-developed leaves were taken from each bush at five-leaf stage and wrapped up on a thin aluminium foil and put on the liquid nitrogen container. After transfixing the samples, they were powdered together with liquid nitrogen in proclaim pounder and 5 mg were poured into the 2 ml tubes and kept in ŌłÆ80┬░C. The genomic DNA was extracted by cetyl trimethyl ammonium bromide (CTAB) as reported by Saghai-Maroof et al. (1984). Quantification and qualification of the extracted DNA was determined by electrophoresis (97 v for 45 min) on 1% agarose gel and DNA samples were diluted about 20-30 ng/╬╝l. The characteristics of 31 SSR markers (Table 2) were extracted from soybase database (www.soybase.org). Polymerase chain reaction (PCR) was carried out using the Eppendorf thermocycler in volume of 15 ╬╝l including: 2 ╬╝l genomic DNA, 1.5 ╬╝l PCR buffer (10├Ś), 0.5 ╬╝l dNTPs (1 mM), 1 ╬╝l of each forward and reverse primers, 1.2 ╬╝l magnesium chloride (15 mM), 0.1 ╬╝l Tag DNA polymerase and 7.7 ╬╝l sterilized distilled water. Thermal cycles were including: one cycle for initial denaturing stage in 95┬░C for 5 min followed by 35 thermal cycles as denaturation in 94┬░C for 30 s, annealing in 45-60┬░C (based on the optimum temperature of each primer) for 30 s and primer extension in 72┬░C for 45 sec and finally after the end of the 35 three-stages thermal cycles, one cycle for final extension in 72┬░C for 5 min. The PCR products were separted by horizontal agarose gel elctrophoresis and the gels were stained by AgNO3 and finally the observed bands for each of the studied genotypes were scored.
To perform the association mapping, structure analysis was firstly conducted to construct the genetic structure matrix of the studied genotypes using the STRUCTURE software (Pritchard et al., 2000). Since there was no prior information on population structure, so the optimum number of groups or sub-populations (K) was determined by simulation, so that K was considered from 1 to 10 and simulation was conducted with period length of 100000 burn in and 100000 Markov Chain Monte Carlo (MCMC) repetition and the optimum K was determined using Evanno et al. (2005) method. Then, membership percentage of each genotype in each group was determined by Spataro et al. (2011) method. Based on this method, a genotype is attributed to a group when its membership percentage is more than 70% (0.7), but if the membership percentage is obtained less than 69% (0.69), it is considered as a mixed genotype. Data obtained from the population structure (Q matrix) and breaking it into two or more sub-populations, was extracted from this software. Finally, the association mapping was conducted using two different statistical models, GLM and MLM, with the data set of phenotypic matrix, genotypic matrix, structure matrix (Q) and kinship matrix by TASSEL 4.1.2 software (Bradbury et al., 2007).
Descriptive statistics of the measured traits including minimum, maximum, mean, range and cofficient of variation (CV) for the studied population is shown in Table 3. The highest CV was observed for grain yield and amount of charcoal rot disease (23.12% and 21.82%, respectively). Furthermore, CV for the other traits was also more than 10%. These results shows that the studied soybean germplasm has a high diversity for the all measured traits that can be useful for the association analysis. Because in the association analysis, genetic factors related to phenotypic variations are searched in more diverse populations than those derived from the crossing of two pure lines, the occurrence of recombinant events during the evolutionary history of these highly diverse populations, which are usually several generations more further from their common ancestry, cause to the failure of the linkage disequilibrium blocks in the genome (Oraguzie et al., 2007). In the other words, all the meiosis events accumulated over the evolutionary history of the plant are considered in the association mapping, while in the conventional mapping populations, the meiosis events occurs only in a few intercourse generations or self-pollination (Oraguzie et al., 2007). Therefore, it seems that to be necessary the existence of a high variation in the studied populations for clarity and accuracy of the results (Oraguzie et al., 2007). This variation was observed in the studied population in this research. Wang et al. (2006) and Vikram (2007) in similar studies in soybean reported the high levels of diversity for their studied populations.
In genetic studies, the population structure which is used to explain the relationships of individuals within and between the populations, provides a perspective on the evolutionary relationships of individuals in a population. Moreover, in the ideal association analysis, there should be no structure in the studied population, indeed, the population should not be structurally divided into subgroups, since the existence of the structure in the studied population could be a deterrent to achieve the reliable results. If the effects of population structure and kinship relationships are not considered in the association analysis, false positive results will arise (Breseghello and Sorrells, 2006). Therefore, understanding the population structure as a prerequisite in association mapping can avoid false positive relationships between markers and traits (Pritchard et al., 2000). In this research, the genetic structure of the studied population and the proper number of sub-population were used as covariate in conducting association analysis based on the Bayesian method in STRUCTURE software (Porras-Hurtado et al., 2013). The results showed that there are three probable sub-populations (K = 3) in the studied germplasm (Fig. 1), which was considered as the optimal K for estimating the population structure and the membership matrix in each sub-population (Q matrix). The results indicated that (Fig. 2) among the total of 130 studied genotypes, 9 genotypes (6.92%) had the mixed structure (the membership probability of each sub-population is less than 0.69), 34 genotypes (26.15%) belonged to the first structure (red), 43 genotypes (33.07%) to the second structure (blue) and 44 genotypes (33.84%) to the third structure (green).
In assoction mapping where the quantitative trait loci (QTLs) are mapped based on the linkage disequilibrium (LD) in addition to combining the population structure, the extent of LD in the genome is also very important (Al-Maskri et al., 2012). In this study, 25.8% of the markers had a significant R2 and greater than 0.1 (R2 Ōēź 0.1, P-value Ōēż 0.01) (Fig. 3). The linkage disequilibrium in the studied genetic population allows association mapping analysis. The factors increasing the amount of LD are system of autogamy, epistasis, genomic alterations, genetic drift, genetic isolation, population structure, small size of population, selection and degree of kinship, while alternating (allogamy), gene transformation, high levels of recombinant and mutation, as well as periodic mutations, are factors that decrease the LD levels (Al-Maskri et al., 2012; Gupta et al., 2005). Slatkin (1999) reported that the multiallelic markers (such as microsatellite) are more likely to achieve a meaningful LD than the bi-allelic markers (such as DArT, SNP, etc.). Remington et al. (2001) also observed a relatively higher range of LDs between SSR markers than SNP markers.
To identify the linked markers to evaluated traits in the studied soybean genotypes, the association mapping was performed based on the general linear model (GLM) dependent on the Q matrix (the membership probability of each individual to each subgroup) and the mixed linear model (MLM) dependent on Q + K matrix (K: kinship relationship matrix) using TASSEL software ver. 3. Based on the results of the GLM, 31 markers showed a significant relationship with the evaluated traits, of which 14 relationships were significant at the probability level of 5% and the others were significant at the probability level of 1%. The associated and significant markers in GLM method were including the relationships between 2 markers with severity of disease, 3 markers with grain yield, 4 markers with each of the traits of number of microsclerota in stem and the amount of charcoal rot disease, 5 markers with pod weight, 6 markers with grain weight and 7 markers with 100 grain weight (Table 4). In contrast, in the MLM model, which uses more information than the GLM model, 35 significant relationships were identified among the studied markers and traits at the probability levels of 5% and 1%, including the relationships of 3 markers with the grain yield, 4 markers with severity of disease, 5 markers with pod weight, number of microsclerota in stem and amount of charcoal rot disease, 6 markers with grain weight and 7 markers with 100 grain weight (Table 4).
A number of common markers were also identified for different traits with both GLM and MLM models in this study. For example, the SSR marker Sat_252 had significant relationships with pod weight, 100 grain weight and amount of charcoal rot disease, Sat_238 with pod weight, grain weight and grain yield, Satt512 with pod weight and 100 grain weight, S63880-CB with grain weight and grain yield, Satt079 with grain weight and 100 seeds weight, Sat_124 with grain weight, 100 seeds weight and grain yield, Satt359, Satt190 and Sat_169 with number of microsclerota in stem, amount of charcoal rot disease and severity of charcoal rot disease, Sat_416 with number of microsclerota in stem and amount of charcoal rot disease and Satt460 with traits of number of microsclerota in stem and amount of charcoal rot disease. This result can probably be due to the pleiotropic effects or tight linkage of the genomic regions involved in controlling these traits (Jun et al., 2008). The identification of common markers is very important in plant breeding since the simultaneous selection of several traits is possible (Tuberosa et al., 2002). Moreover, the significant relationships between several markers with a specific trait was also showed in this research (Table 4). For example, the relationship between Sat_404 and Satt361 markers with the pod weight, Sct_028 and Satt460 markers with grain weight and Satt607, Sat_357 and Satt644 markers with 100 grain weight, indicating the quantitative and polygenic inherentance of the evaluated traits. On the other hand, low values of the coefficients of determination (R2) for most of the linked markers also confirms the same issue and shows the determination of some variances in these traits through identified genetic locations and, therefore, the greater effect of the environment (relative to genetic effects) on variation of these traits. In general, considering the constraints of the linkage mapping method, such as the lack of availability of dispersed populations and the long time required to create them, the association analysis method by eliminating these limitations provides researchers with appropriate marker information (Oraguzie et al., 2007).
The results of the present study showed the effectiveness of the association mapping method in identifying markers linked to the evaluated traits in the studied soybean genotypes. Evidently, it is necessary to validate the markers identified and associated with relevant traits in large populations with a higher level of diversity as well as in different environments, in order to ensure their relevance to the related traits, and thus to increase the efficiency these markers will increase in various breeding programs such as marker assisted selection (MAS). Neale and Savolainen, (2004) showed that genetic locations selected by the association analysis have important advantages such as involving adequate levels of nucleotide diversity and also the ability accurately phenotypic evaluations that can be used in MAS. Several studies have previously been conducted to identify genetic locations associated with resistance to charcoal rot disease in different plants by different molecular markers. Olaya et al. (1996) showed that the resistance to charcoal rot disease in soybean was controlled by two genes with complete dominance, called mp-1 and mp-2. They also identified two RAPD markers related to resistance. Miklas et al. (1998) identified three QTLs associated with resistance to charcoal rot disease in beans by association mapping. Yuan et al. (2002) showed that Satt294 marker on C1 linkage group, Satt440 on I linkage group and Satt337 on K linkage group are associated with seed yield in soybean. Hern├Īndez-Delgado et al. (2009) showed that the resistance to charcoal rot in beans is controlled by two dominant genes with epistatic effects. Sun et al. (2014) by association analysis identified four SSR alleles, Satt 634-133, Satt 634-149, Satt 222-168 and Satt 301-190, which were associated with a slight resistance to phytophthora disease in soybean.
Acknowledgments
We thanks from the University of Guilan, Rasht, Iran, and the Seed and Plant Improvement Research Institute (SPIRI), Karaj, Iran, for their financial support of this research.
Fig.┬Ā1
Bilateral charts for determining the number of sub-populations in the studied soybean genotypes (K=3) based on microsatellite markers.
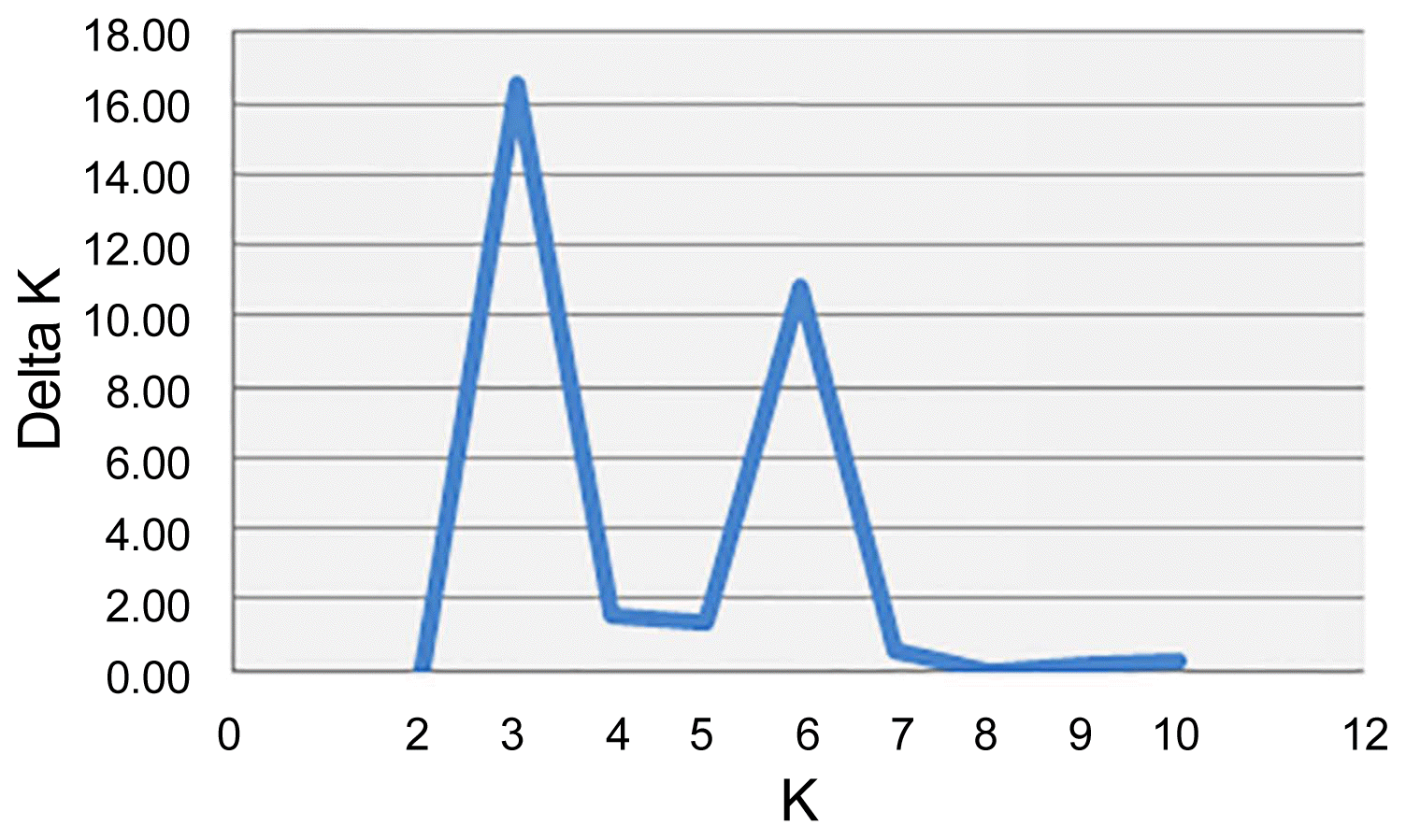
Fig.┬Ā2
Bayesian model based cluster analysis for 130 different soybean genotypes using 31 microsatellite loci (K=3). Each color indicate one sub-population or cluster. Vertical axis show the membership coefficient of each genotype into clusters.
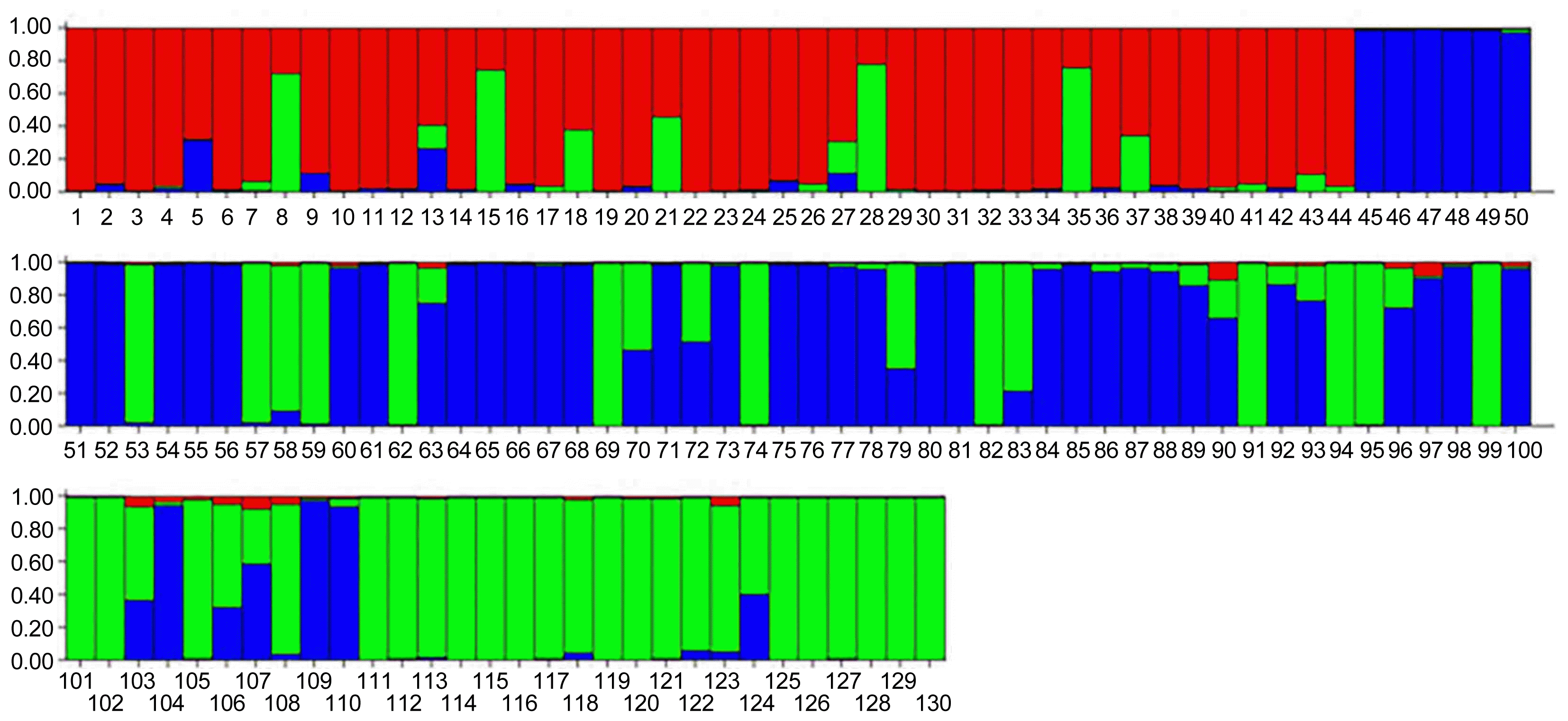
Fig.┬Ā3
Linkage disequilibrium plot (LD plot). Diameter upper and lower are indicating linkage disequilibrium and p-value for each pair of marker, respectively.

Table┬Ā1
Soybean genotypes evaluated to charcoal rot disease (Macrophomina phaseolina) in field conditions in this research
| No. | Genotype | Maturity groupa | Codeb | No. | Genotype | Maturity groupa | Codeb | |
|---|---|---|---|---|---|---|---|---|
| 1 | AGS 358 (3) | II | 2176 | 33 | Si-bi-va- 1207 | II | 2038 | |
| 2 | AGS 359 (4) | III | 3072 | 34 | A 3237 | II | 2019 | |
| 3 | Hartwig | III | 3061 | 35 | A 3935 | II | 2020 | |
| 4 | Gloy | III | 3062 | 36 | Columbus | II | 2022 | |
| 5 | 2-L.80-5914 | III | 3057 | 37 | Union | II | 2034 | |
| 6 | B-R22 Bijelina | I | 1173 | 38 | Stressland-B | III | 3034 | |
| 7 | LN 89-3394 | II | 2149 | 39 | Stressland-C | III | 2043 | |
| 8 | L.D 9 | II | 2079 | 40 | GN3074 | III | 3074 | |
| 9 | Kenwood | II | 2099 | 41 | Pek - Cak - taj | III | 3031 | |
| 10 | Fowler | III | 3045 | 42 | Swift | III | 3026 | |
| 11 | TN 4.94 | III | 3037 | 43 | G.3├Ś Hamilton (10) | V | Sh8 | |
| 12 | Manacon | III | 3022 | 44 | DPX ├Ś Yougetsu (2) | V | Sh18 | |
| 13 | Fowler | III | 3045 | 45 | DPX ├Ś Yougetsu (3) | V | Sh19 | |
| 14 | Cysne | II | 2093 | 46 | DPX ├Ś Darby (2) | V | Sh31 | |
| 15 | Sort 62 | II | 2064 | 47 | DPX ├Ś Darby (3) | V | Sh32 | |
| 16 | Sort 126 S.M.A.B | II | 2065 | 48 | Williams ├Ś DPX (6) | V | Sh40 | |
| 17 | Wars zawska | II | 2044 | 49 | Hamilton ├Ś Sahar (3) | V | Sh47 | |
| 18 | Bonus | II | 2041 | 50 | Hamilton ├Ś Nemaha (6) | V | Sh55 | |
| 19 | Clean | II | 2052 | 51 | 9242 | II | 2004 | |
| 20 | Stressland-A | II | 2055 | 52 | S 24 - 92 | II | 2005 | |
| 21 | 5601-46-6-1 C | II | 2056 | 53 | CX 232 | II | 2006 | |
| 22 | Harbinskaia 111-3994/56 | II | 2060 | 54 | Karbine | I | 1096 | |
| 23 | Bean - Comet B | II | 2061 | 55 | Harbinskaia 3971 B | I | 1097 | |
| 24 | Delsoy 4210 | III | 3017 | 56 | Dikmanova - Cierna | I | 1098 | |
| 25 | Comet (NRM) B | I | 1160 | 57 | Dornburger | I | 1099 | |
| 26 | B-R23 Bijelina | I | 1162 | 58 | Banjaluka B | II | 1100 | |
| 27 | Bijelina 54/68 | I | 1163 | 59 | Harasoy | I | 1090 | |
| 28 | NS-16 B | I | 1139 | 60 | Motte | IV | 4001 | |
| 29 | B-R3 (Bijelina) | I | 1140 | 61 | K.S 4895 | IV | 4007 | |
| 30 | Grangelb | I | 1118 | 62 | Essex | IV | 4009 | |
| 31 | Mishel | II | 2042 | 63 | AGS 381 (10) | IV | 4010 | |
| 32 | Calland | II | 2047 | 64 | TN 5.95 | V | 5001 | |
| 65 | Delsoy 4710 | V | 5002 | 98 | NE-3297 | II | 2133 | |
| 66 | EJC (Edi. Jappan) | V | 5003 | 99 | ST.Pazova 54/18 | II | 2162 | |
| 67 | Telar | V | 5004 | 100 | N.S-L-11-6 | II | 2161 | |
| 68 | Nekador | V | 5005 | 101 | Mercory | II | 2147 | |
| 69 | Hatcheson | V | 5006 | 102 | Roanak | II | 2125 | |
| 70 | Cliford | V | 5007 | 103 | Pance Vacka B | II | 2123 | |
| 71 | Hood | V | 5008 | 104 | L.52 | II | 2165 | |
| 72 | Kaspian | V | 5009 | 105 | Sort 126 S.M.A.B | II | 2164 | |
| 73 | Sari | V | 5010 | 106 | ERFurt | II | 2163 | |
| 74 | AGS 346 (2) | V | 5011 | 107 | VINIMK 9186 | II | 2117 | |
| 75 | AGS (5) | V | 5012 | 108 | PA 83 | II | 2098 | |
| 76 | AGS 367 (6) | V | 5013 | 109 | VESTAG 97 | II | 2097 | |
| 77 | AGS 364 (8) | V | 5014 | 110 | Hack | II | 2095 | |
| 78 | AGS 380 (9) | V | 5014 | 111 | Hadgson | II | 2027 | |
| 79 | Doles | V | 5018 | 112 | CM - 1070 | II | 2012 | |
| 80 | GN2050 | V | 5020 | 113 | S - 12 - 49 | II | 2013 | |
| 81 | DI 74 | V | 17F-1 | 114 | S.R.F ├Ś Columbus | II | 2016 | |
| 82 | D42.I4 | III | 17F-4 | 115 | Budgoszkasz 061 | II | 2118 | |
| 83 | Linford | III | 17F-13 | 116 | Rounest | II | 2119 | |
| 84 | Clean | III | 17F-14 | 117 | Poplu - 18 - 35 | II | 2028 | |
| 85 | LH-2500 | III | 17F-15 | 118 | Tokyo Brown | II | 2029 | |
| 86 | M 7 | III | 17F-16 | 119 | Century 84 | II | 2030 | |
| 87 | TN 6.90 | III | 2130 | 120 | RCAT ANGORA | II | 2007 | |
| 88 | T 215 | II | 2171 | 121 | S19 - 90 | II | 2009 | |
| 89 | Kabalovskaja B | II | 2167 | 122 | Black Tokyo | II | 2062 | |
| 90 | Kabalovskaja | II | 2166 | 123 | Cul.9132 | I | 1047 | |
| 91 | 8-L.65-3266 | II | 2157 | 124 | AP - 1394 | I | 1098 | |
| 92 | Black Hawck | II | 2156 | 125 | PRO - 280 | I | 1064 | |
| 93 | Illinoi | II | 2155 | 126 | S 14 - H 4 | I | 1065 | |
| 94 | L.2 | II | 2152 | 127 | SENTRY | I | 1066 | |
| 95 | S3-941-8-1-8 | II | 2142 | 128 | Spirit | II | TU38 | |
| 96 | L.8 | II | 2140 | 129 | Salin | II | TU309 | |
| 97 | Darby | II | 2138 | 130 | Interprise | II | 1067 | |
Table┬Ā2
Characteristics of the studied microsatellite markers in this research
Table┬Ā3
Minimum, maximum, mean ┬▒ standard deviation (SD), range and phenotypic coefficient of variation of the measured traits in the 130 soybean genotypes studied in this research
Table┬Ā4
Microsatellite markers linked to evaluated traits in the studied soybean population using the association mapping based on GLM and MLM models
References
Babu, BK, Saxena, AK, Srivastava, AK and Arora, DK 2007. Identification and detection of Macrophomina phaseolina by using specific species oligonucleotide primers and prob. Mycologia. 99:797-803.


Bilyeu, KD, Ratnaparkhe, MB and Kole, C 2010. Genetics, genomics and breeding of soybean. 1st ed. CRC Press, Boca Raton USA. 388.
Bradbury, PJ, Zhang, Z, Kroon, DE, Casstevens, TM, Ramdoss, Y and Buckler, ES 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 23:2633-2635.


Breseghello, F and Sorrells, ME 2006. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 172:1165-1177.



Coser, SM, Chowda Reddy, RV, Zhang, J, Mueller, DS, Mengistu, A, Wise, KA, Allen, TW, Singh, A and Singh, AK 2017. Genetic architecture of charcoal rot (Macrophomina phaseolina) resistance in soybean revealed using a diverse panel. Front Plant Sci. 8:1626



Evanno, G, Regnaut, E and Goudet, E 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14:2611-2620.


Fusari, CM, Di Rienzo, JA, Troglia, C, Nishinakamasu, V, Moreno, MV, Maringolo, C, Quiroz, F, Alvarez, D, Escande, A, Hopp, E, Heinz, R, Lia, VV and Paniego, NB 2012. Association mapping in sunflower for Sclerotinia head rot resistance. BMC Plant Biol. 12:93



Hern├Īndez-Delgado, S, Reyes-Vald├®s, MH, Rosales-Serna, R and Mayek-Perez, N 2009. Molecular markers associated with resistance to Macrophomina phaseolina (Tassi) Goid in common bean. J Plant Pathol. 91:163-170.
Iqbal, MJ, Meksem, K, Njiti, VN, Kassem, A and Lightfoot, DA 2001. Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden-death syndrome (SDS) in Essex ├Ś Forrest RILs. Theor Appl Genet. 102:187-192.

Iquira, E, Humira, S and Francois, B 2015. Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol. 15:5



Jana, T, Sharma, TR, Prasad, RD and Arora, DK 2003. Molecular characterization of Macrophomina phaseolina and Fusarium species by a single primer RAPD technique. Microbiol Res. 158:249-257.


Li, XP, Han, YP, Teng, WL, Zhang, SZ, Yu, KF, Poysa, V, Anderson, T, Ding, J and Li, W 2010. Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ConradŌĆÖ and Hefeng 25. Theor Appl Genet. 121:651-658.


Porras-Hurtado, L, Ruiz, Y, Santos, C, Phillips, C, Carracedo, A and Lareu, MV 2013. An overview of STRUCTURE: applications, parameter settings and supporting software. Front Genet. 4:98



Mengistu, A, Ray, JD, Smith, JR and Paris, RL 2007. Charcoal rot disease assessment of soybean genotypes using a colony forming unit index. Crop Sci. 47:2453-2461.

Mengistu, A, Arelli, PA, Bond, JP, Shannon, GJ, Wrather, AJ, Rupe, JB, Chen, P, Little, CR, Canaday, CH, Newman, MA and Pantalone, VR 2011. Evaluation of soybean genotypes for resistance to charcoal rot. Plant Health Prog. 12:44-59.

Musial, JM, Mackie, JM, Armour, DJ, Phan, HTT, Ellwood, SE, Aitken, KS and Irwin, JAG 2008. Identify of QTL for resistance and susceptibility to Stagonospora meliloti in autotetraploid lucern. Theor Appl Genet. 1148:1427-1435.
Neale, DB and Savolainen, O 2004. Association genetic of complex traits in conifers. Trends Plant Sci. 9:325-330.

Olaya, G, Abawi, GS and Weeden, NF 1996. Inheritance of the resistance to macrophomina phaseolina and identification of RAPD markers linked to resistance genes in beans. Phytopathology. 86:674-679.

Oraguzie, NC, Rikkerink, EHA, Gardine, SE and de Silva, HN 2007. Association mapping in plants. Springer, NY, USA. 278.
Pritchard, JK, Stephens, M and Donnelly, P 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945-959.




Remington, DL, Thornsberry, JM, Matsuoka, Y, Wilson, LM, Whitt, SR, Doebley, J, Kresovich, S, Goodman, MM and Buckler, ES 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA. 98:11479-11484.



Saghai-Maroof, MA, Soliman, KM, Jorgensen, RA and Allard, RW 1984. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA. 81:8014-8018.



Singh, RJ 2006. Genetic resources, chromosome engineering and crop improvement: Oilseed crops. 3rd ed. CRC Press, Boca Raton, FL USA. 320.
Spataro, G, Tiranti, B, Arcaleni, P, Bellucci, E, Attene, G, Papa, R, Spagnoletti Zeuli, P and Negri, V 2011. Genetic diversity and structure of a worldwide collection of Phaseolus coccineus L. Theor Appl Genet. 122:1281-1291.


Sun, J, Guo, N, Lei, J, Li, L, Hu, G and Xing, H 2014. Association mapping for partial resistance to Phytophthora sojae in soybean (Glycine max (L.) Merr.). J Genet. 93:355-363.


Tesso, T and Ejeta, G 2011. Stalk strength and reaction to infection by Macrophomina phaseolina of brown midrib maize (Zea mays) and (Sorghum bicolor). Field Crops Res. 120:271-275.

Tuberosa, R, Gill, BS and Quarrie, SA 2002. Cereal genomics: Ushering in a brave new world. Plant Mol Biol. 48:445-449.

Vikram, D 2007. Diversity analysis in soybean [Glycine max (L.) Merrill] using morphological and simple sequence repeat (SSR) markers. MasterŌĆÖs thesis. Tamil Nadu Agricultural University, Coimbatore, India.
Wang, L, Guan, R, Zhangxiong, L, Chang, R and Qiu, L 2006. Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci. 46:1032-1038.

Wang, J, McClean, PE, Lee, R, Goos, RJ and Helms, T 2008. Association mapping of iron deficiency chlorosis loci in soybean [Glycine max (L.) Merr.] advanced breeding lines. Theor Appl Genet. 116:777-787.


- TOOLS
-
METRICS

- Related articles
-
Message in a Bottle: Chemical Biology of Induced Disease Resistance in Plants2008 September;24(3)
First Description of Crown Gall Disease on Ginseng2008 June;24(2)
Classification and Function of Plant Disease Resistance Genes1999 April;15(2)
Pseudomonas avenae Causing Bacterial Brown Stripe Disease of Rice in Korea1985 April;1(1)



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



