Fusarium Species from Sorghum in Thailand
Article information
Abstract
Sorghum is the fifth most important cereal worldwide, spreading from Africa throughout the world. It is particularly important in the semi-arid tropics due to its drought tolerance, and when cultivated in Southeast Asia commonly occurs as a second crop during the dry season. We recovered Fusarium from sorghum in Thailand and found F. proliferatum, F. thapsinum and F. verticillioides most frequently, and intermittent isolates of F. sacchari and F. beomiforme. The relatively high frequencies of F. proliferatum and F. verticillioides, suggest mycotoxin contamination, particularly fumonisins and moniliformin, should be evaluated. Genetic variation within the three commonly recovered species was characterized with vegetative compatibility, mating type, Amplified Fragment Length Polymorphisms (AFLPs), and female fertility. Effective population number (Ne) was highest for F. verticillioides and lowest for F. thapsinum with values based on mating type allele frequencies higher than those based on female fertility. Based on AFLP genetic variation, the F. thapsinum populations were the most closely related, the F. verticillioides populations were the most distantly related, and the F. proliferatum populations were in an intermediate position. The genetic variation observed could result if F. thapsinum is introduced primarily with seed, while F. proliferatum and F. verticillioides could arrive with seed or be carried over from previous crops, e.g., rice or maize, which sorghum is following. Confirmation of species transmission patterns is needed to understand the agricultural systems in which sorghum is grown in Southeast Asia, which are quite different from the systems found in Africa, Australia, India and the Americas.
Sorghum (Sorghum bicolor) is an important crop for human consumption, animal feed, and bioenergy, and the fifth most important cereal in the world after wheat, rice, maize, and barley. From its domestic origins in Africa, the crop has spread throughout the world and now is cultivated on all inhabited continents. In Thailand, sorghum is grown in all regions of the country, but most commonly in the central and northeastern regions. Much of the sorghum grown in Thailand is exported, although some is used as animal feed and for ethanol production (Ariyajaroenwong et al., 2012; Boon Long, 1992; Nuanpeng et al., 2011). Sorghum is the third most important cereal crop in Thailand (after rice and maize), and the country ranks 18th in the world for sorghum exports (http://www.indexmundi.com/agriculture/). The average production of sorghum from 2006 to 2012 was 54,500 tons per year which has decreased by more than 50% compared to production in 2000 (http://faostat.fao.org). China and India are the two largest sorghum producers in Asia, with production occurring under conditions similar to those in Africa and the Americas. In Southeast Asia, sorghum usually is cultivated as a second crop during the dry season.
Fusarium spp. from sorghum have been poorly differentiated and described in Southeast Asia, although there has been great progress in identifying and differentiating Fusarium species from this host over the last 20–30 years. In many parts of the world, the only Fusarium species reported to occur on sorghum is Fusarium moniliforme. This name was retired in 2003 (Seifert et al., 2003), as it now refers to fungi in 15–50 different species. Fusarium moniliforme is the only Fusarium species that has been reported to occur on sorghum in Thailand (Boon Long, 1992; Salleh et al., 1995), although many other Fusarium species are known to occur there.
Some of the most important diseases of sorghum are stalk rot and grain mold, both of which have a Fusarium causal agent (Adeyanju et al., 2015; Bandyopadhyay et al., 2000; Choi et al., 2013; Funnell-Harris et al., 2016; Leslie, 2002; Little et al., 2012; Marasas et al., 2001; Upadhyaya, et al., 2013). Stalk rot incidence may be up to 90% and losses up to 50%, although year-to-year losses are usually much less (Jardine, 2006). Numerous additional Fusarium species from sorghum have been identified from sorghum, including F. andiyazi, F. napiforme, F. nygamai, F. proliferatum, F. pseudonygamai, F. thapsinum, and F. verticillioides (Klittich et al., 1997; Leslie and Summerell, 2006; Leslie et al., 1990, 2005a; Marasas et al., 1987, 2001; Nelson et al., 1987). A number of these species had not been formally described prior to the last published reports from Thailand (Pitt et al., 1994; Salleh et al., 1995). Some of these species produce fumonisins, which are toxic to humans and domesticated animals, and other mycotoxins of as yet unconfirmed importance, such as moniliformin, fusaric acid and fusaproliferin (Desjardins, 2006). Sorghum cultivated in Southeast Asia is grown in a very different climate than occurs in most of Africa, the Americas or Australia, where much of the work on sorghum pathology has occurred. The indigenous fungal populations in Southeast Asia differ from those known from other locations where sorghum is cultivated, as does the rice-focused cropping system. The general lack of information on sorghum pathogens in Southeast Asia and the ongoing revisions to the nomenclature of the relevant Fusarium species over the last 20 years necessitate a reevaluation of the Fusarium species from this region to determine if the species recovered from sorghum grown in Southeast Asia are comparable to those recovered from sorghum grown in other better-studied geographic locations.
The objective of this study was to identify the Fusarium species present on cultivated sorghum in Thailand and to evaluate their relatedness. We hypothesize that Fusarium species within the Fusarium fujikuroi species complex associated with sorghum in other parts of the world will be present, although many of these species have not previously been reported from Southeast Asia. Our data can be used to develop or adapt disease management strategies and to estimate risks of mycotoxin contamination of sorghum grain produced in this region.
Materials and Methods
Isolates and culture conditions
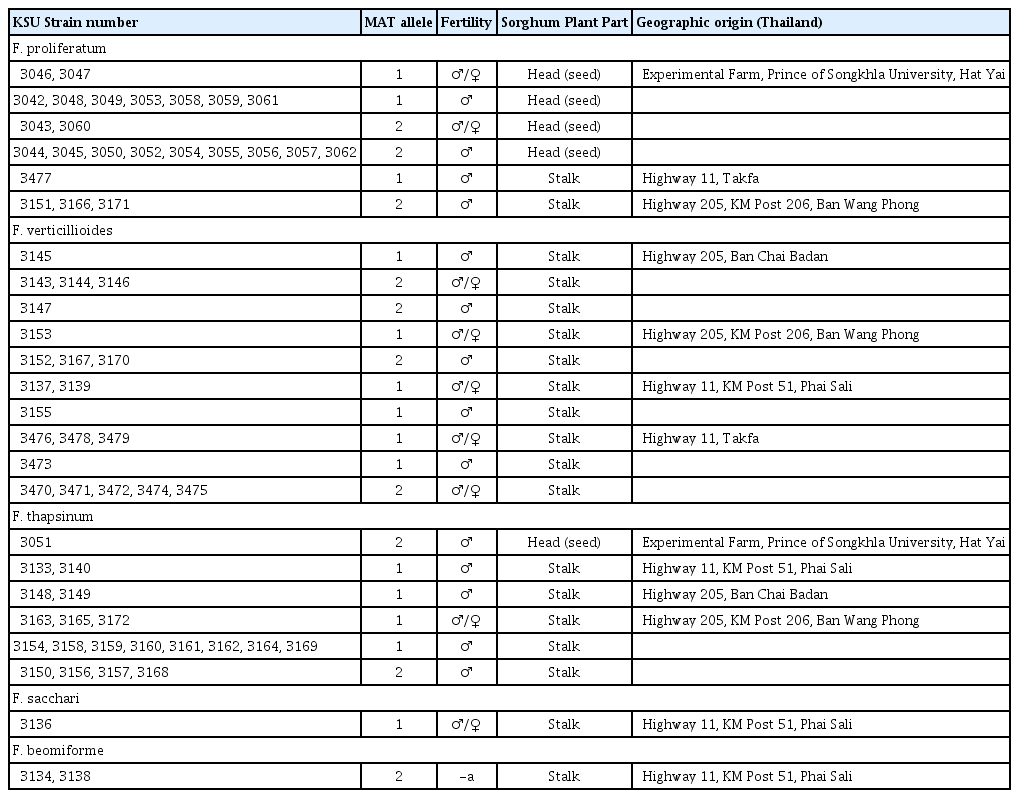
Forty-seven isolates of Fusarium spp. were recovered from stalks and 21 isolates were recovered from seeds of asymptomatic sorghum plants at five locations in Thailand (Table 1). Samples from roots and soil were not collected. Most isolates were recovered from different plants, and only genetically unique isolates were included in the analyses if more than one isolate was analyzed from a plant, i.e., clonal isolates recovered from the same plant were “censored” (Table 2). Isolates were recovered from plant tissue placed on a semi-selective peptone/PCNB medium (Leslie and Summerell, 2006), and purified by subculturing individual microconidia separated by micromanipulation. The resulting cultures were preserved as spore suspensions in 15% glycerol at −70ºC.
Reference strains used in this study were F. verticillioides (FGSC 7415 and FGSC 7416), F. sacchari (FGSC 7419 and FGSC 7420), F. fujikuroi (KSU 1993 and KSU 1994), F. proliferatum (FGSC 7422 and FGSC 7421), F. subglutinans (FGSC 7616 and FGSC 7617), F. thapsinum (FGSC 7056 FGSC 7057). and F. nygamai (KSU 5112), and F. circinatum (FGSC 9022 and FGSC 9023) [FGSC – Fungal Genetics Stock Center; Department of Plant Pathology, Kansas State University, Manhattan, Kansas].
DNA isolation
Isolates were cultured on complete medium (Leslie and Summerell, 2006) slants for seven days. One ml of a spore suspension in 0.25% Tween® 60 solution (~106 spores/ml) was used to inoculate 40 ml of complete medium broth in a 125 ml flask, and then cultured for two days at room temperature (22–26°C) on an orbital shaker (150 rpm). Mycelia were harvested by filtration through a Milk Filter disk (KenAG, Ashland, Ohio, USA), dried by blotting with paper towels and ground to a powder in a mortar with a pestle under liquid nitrogen. DNA was extracted by using a CTAB protocol (Leslie and Summerell, 2006). DNA extracts were stored in 1.5 ml microcentrifuge tubes at 4ºC until used. The quality of DNA was evaluated following separation on a 1% agarose gel. DNA concentrations were measured with a Nanodrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, Delaware, USA).
Mating type PCR
DNA solutions were diluted to ~20 ng/μl with sterile double-distilled water. The MAT alleles were amplified by PCR as previously described (Leslie and Summerell, 2006), with the primers developed by Steenkamp et al. (2000) and Kerényi et al. (1999, 2004). PCR amplification products were resolved on a 1% agarose gel, and the bands present were used to identify the mating type.
Biological species and female fertility
Isolates of F. proliferatum, F. sacchari, F. thapsinum, and F. verticillioides were crossed with standard, female-fertile, tester isolates. Sexual crosses were made on carrot agar as described by (Klittich and Leslie, 1988). Fertility was determined by the presence of perithecia exuding a cirrus of ascospores 2–4 weeks after fertilization. Positive crosses were repeated twice and negative crosses were repeated three times.
Field isolates were tested as both male and female parents in crosses with the standard testers after the MAT allele in the field isolate was identified. Male fertility was scored in crosses in which the field isolate was the male and the standard tester strain was the female parent. Female fertility was scored in crosses in which the roles were reversed and the field isolate was the female parent and the standard tester strain was the male. Estimates of effective population number were made by using the equations of Leslie and Klein (1996).
Vegetative compatibility groups (VCGs)
Vegetative compatibility was assessed by pairing complementary nitrate non-utilizing (nit) mutants that had been generated on chlorate medium (Correll et al., 1987; Leslie and Summerell, 2006). NitM and nit1 mutants that were heterokaryon self-compatible (Correll et al., 1989) were paired preferentially, with nit1 and nit3 mutants paired if a NitM mutant was not available. Mutants that formed prototrophic heterokaryons when paired were considered to be in the same VCG, and those that did not were considered to be in different VCGs.
AFLP reactions and analysis
DNA fingerprints were generated by using AFLPs (Vos et al., 1995) following the protocol described in Leslie and Summerell (2006). Three primer pairs were used in the selective amplification: EcoRI+GG(5′-AGACTGCGTACCAATTCGG-3′)/MseI+CT(5′-GATGAGTCCTGAGTAACT-3′), EcoRI+AA(5′-AGACTGCGTACCAATTCAA-3′)/MseI+TT(5′-GATGAGTCCTGAGTAATT-3′), and EcoRI+TT(5′-AGACTGCGTACCAATTCTT-3′)/MseI+AC(5′-GATGAGTCCTGAGTAAAC-3′). Bands 200–500 bp in length were scored manually based on the presence or absence of a band. Individual bands were assumed to represent an allele at a single locus. Similarities between all strains were analyzed by the neighbor-joining clustering option of PAUP (version 4.10b; D. L. Swofford, Sinauer Associates, Sunderland, Massachusetts) with 1000 bootstrapping replications. Estimates of molecular variance, based on the AFLP genotypes, within and among populations were calculated by using analysis of molecular variance (AMOVA) as implemented in GenAlEx 6.5 (Peakall and Smouse, 2012).
DNA sequencing
Partial sequences of the translation elongation factor 1α (tef-1) gene were analyzed for selected isolates in each major cluster identified by the AFLP analysis. DNA samples for sequencing were diluted to ~20 ng/μl. The primer sequence used for tef-1 was EF-1 (forward: 5′-ATGGGTAAGGAGGACAAGAC-3′) and EF-2 (reverse: 5′-GGAAGTACCAGTGATCATGTT-3′) (O’Donnell et al., 1998). The conditions for tef-1 amplification were: 94ºC for 1 min., followed by 34 cycles of 94ºC for 30 sec., 62ºC for 45 sec., and 72ºC for 1 min., and then held at 4ºC indefinitely. The amplification products for tef-1 were cleaned with ExoSAP-IT (Affymetrix, Cleveland, OH) following the manufacturer’s protocol. DNA sequences were obtained by using an Applied Biosystems 3730 DNA Analyzer at the K-State Sequencing facility. These DNA sequences were analyzed with BioEdit (Hall, 1999) and BLASTed against the NCBI GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and a Fusarium-specific database (http://isolate.fusariumdb.org/index.php; Geiser et al., 2004).
Results
Identification and species distribution
All 68 isolates recovered could be assigned to one of five species (Table 1): F. proliferatum, F. verticillioides, F. thapsinum, F. beomiforme and F. sacchari. Species identity was based on at least two of: AFLP analysis, partial gene sequence of tef-1, and cross fertility with standard tester strains. Three species were common – F. proliferatum (35%), F. verticillioides (31%), and F. thapsinum (29%) (Table 3). Fusarium proliferatum and F. thapsinum were the only species recovered from sorghum heads (seeds) (Table 1), although all five species were recovered from stalks. Fusarium proliferatum dominated numerically in samples from sorghum heads, but was found at only three of the five locations. Fusarium thapsinum and F. verticillioides were more widely distributed, being recovered at four locations (Table 1), but only F. thapsinum was recovered from sorghum heads. Fusarium beomiforme and F. subglutinans were both recovered from only a single site – Phai Sali (Table 1). Four species were recovered from this site, while samples from the other sites yielded either two or three species per site.
AFLP analyses
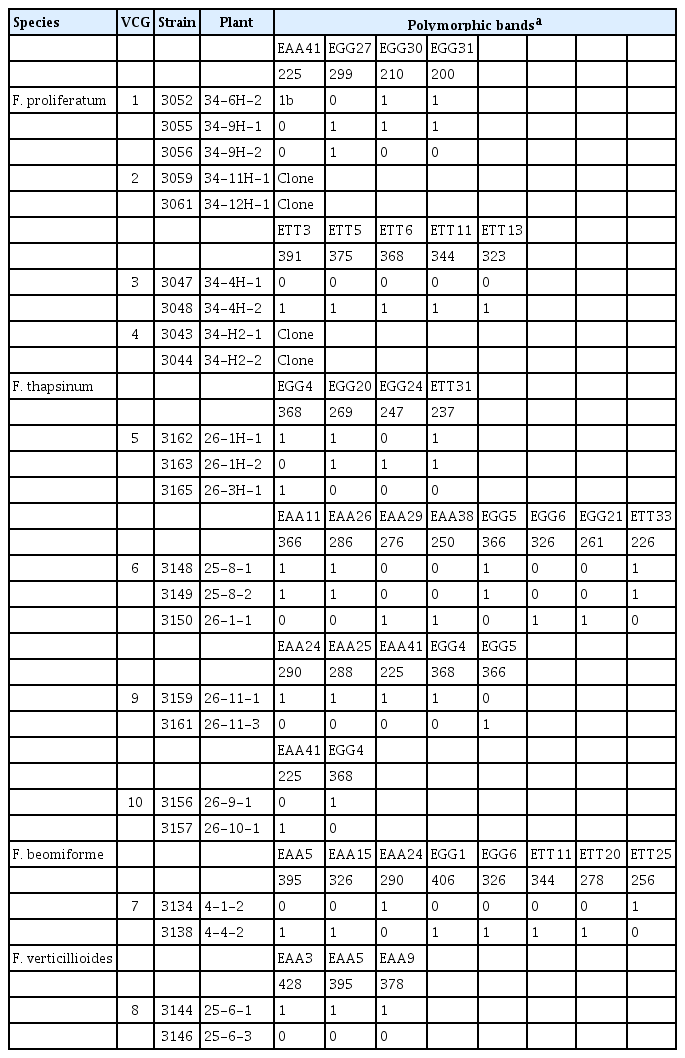
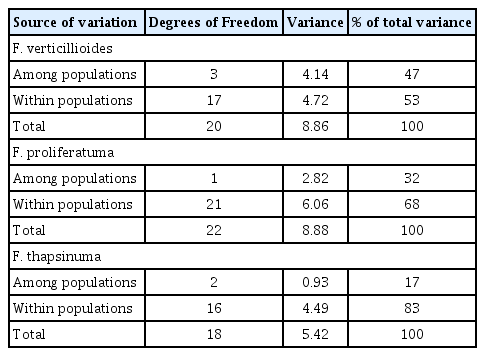
Seventy-eight polymorphic AFLP bands were scored manually as the presence or absence of the band. All of the isolates from the same species clustered in unique clades in an unrooted tree (Fig. 1). Reference strains were included in this analysis. Identity of isolates that did not cluster with a reference strain, e.g., F. beomiforme, was confirmed with partial tef-1 gene sequences. Analysis of molecular variance (AMOVA) based on the AFLP bands was calculated for F. verticillioides, F. proliferatum, and F. thapsinum (Table 4). Genetic variation within F. proliferatum, F. thapsinum and F. verticillioides was 68%, 83% and 53%, respectively.
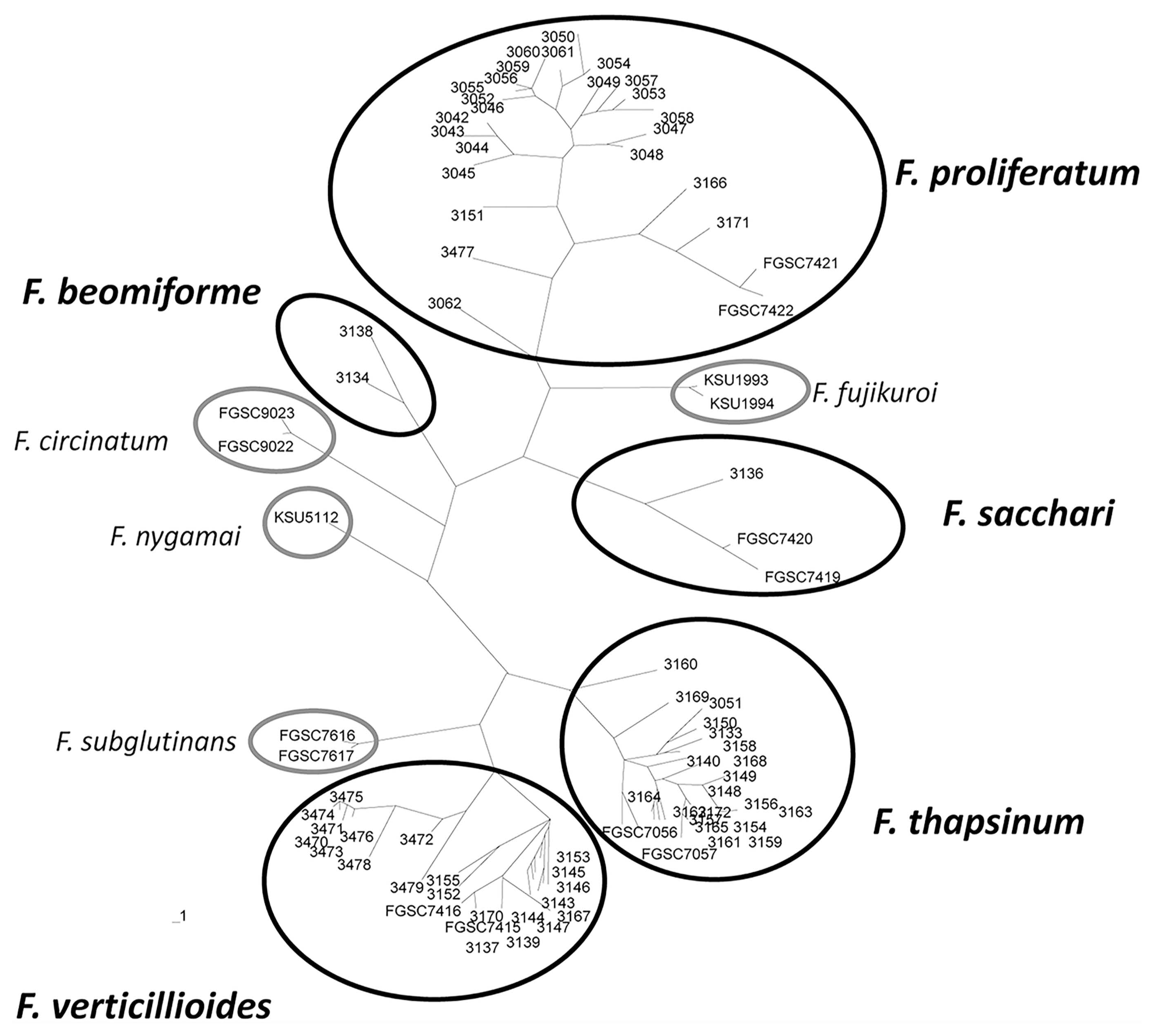
Unrooted phylogenetic tree generated with UPGMA based on AFLP markers (Page, 1996). Solid black circles encompass strains from species identified from the field populations. Light gray circles encompass reference strains for related species. Strains deposited in the Fungal Genetics Stock Center are preceded by “FGSC”. Other strains are all from the authors’ collection at Kansas State University.
Vegetative compatibility
One strain of F. proliferatum was heterokaryon self-incompatible and was excluded from this study. The remaining strains were all heterokaryon self-compatible. Ten multi-member VCGs were found amongst the strains evaluated – four each within F. proliferatum and F. thapsinum, and one each within F. beomiforme and F. verticillioides – and contained 23 strains (Table 2). Strains not in one of these 10 multi-member groups were all in single-member VCGs. One of the VCGs from F. proliferatum and two of the VCGs from F. thapsinum contained three isolates each, while the seven other multi-member VCGs contained only two strains each.
Strains within a VCG could be, but were not necessarily, genetic clones, i.e., did not have the same AFLP haplotype. In two of the F. proliferatum VCGs, both of the isolates in the VCG had the same AFLP haplotype. In one of the three-membered F. thapsinum VCGs, two of the strains had the same AFLP haplotype and differed from the third member of the VCG at eight of the AFLP loci. Strains with the same AFLP haplotype could both be recovered from the same plant, and could represent re-isolations of the same strain. In seven VCGs, members of the same VCG had different AFLP haplotypes and differed from one another at two to four loci in the remaining two F. proliferatum VCGs, at two to five loci in the three remaining F. thapsinum VCGs, at eight loci in the F. beomiforme VCG, and at three loci in the F. verticillioides VCG.
Twenty-four total AFLP loci varied amongst the strains within the multi-member VCGs, with 20 varying within only a single VCG. The remaining four AFLP loci were polymorphic within multiple VCGs. There was no particular pattern to the VCGs in which these loci varied – one varied in a single VCG of F. proliferatum and two VCGs of F. thapsinum, a second in a VCG of F. proliferatum and a VCG of F. thapsinum, a third in a VCG of F. proliferatum and the VCG of F. beomiforme, and the fourth in the VCG of F. beomiforme and the VCG of F. verticillioides. The VCG of F. beomiforme was the only VCG in which two of the four loci that varied in more than one VCG were found.
Mating type and female fertility
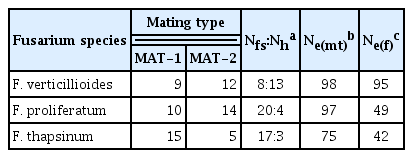
Mating type and female fertility were scored for strains in all species except F. beomiforme, which lacks a known sexual stage (Table 3). Both isolates of F. beomiforme were MAT-2. The MAT alleles in F. proliferatum and F. verticillioides were both present in roughly equal frequencies. In F. thapsinum, there were significantly (χ2 = 5.0; P ≤ 0.05) more strains with the MAT-1 genotype than with the MAT-2 genotype. The only isolate of F. subglutinans was MAT-1.
Female fertility in F. verticillioides was relatively high, with 62% of the strains being female fertile, while 38% were female sterile. Amongst strains of F. proliferatum and F. thapsinum, female fertility was low, at 17% and 15%, respectively.
Discussion
All five species we recovered from Thailand have previously been associated with sorghum from elsewhere in the world, and all but F. beomiforme are well-documented as sorghum pathogens. Four of these species (F. proliferatum, F. thapsinum, F. verticillioides, and F. sacchari) are from the Fusarium fujikuroi species complex (also known broadly as the Gibberella fujikuroi species complex or Fusarium section Liseola). Fusarium beomiforme shares many morphological features with the other four species, but is phylogenetically relatively distant from them, as it is a member of the F. burgessii species complex (Laurence et al., 2011, 2015). The three species we recovered most frequently, F. proliferatum, F. verticillioides, and F. thapsinum, are commonly associated with sorghum (Leslie, 2002; Leslie and Summerell, 2006; Leslie et al., 1990; Lincy et al., 2011; Onyike and Nelson, 1992; Sharma et al., 2011; Tarekegn et al., 2006). Fusarium sacchari is less commonly recovered from sorghum, and often is associated with a leaf spot (Leslie et al., 2005b). Previous reports of F. subglutinans associated with sorghum also may be attributable to F. sacchari, since these two species are difficult to distinguish based solely on morphology (Leslie and Summerell, 2006) and were recognized as members of different biological species only in 1991 (Leslie, 1991; Leslie et al., 2005b). Other Fusarium species previously recovered from sorghum include F. andiyazi (Marasas et al., 2001), F. graminearum (Choi et al., 2013; Menkir et al., 1996; Quazi et al., 2010; Tarekegn et al., 2006), F. nygamai (del Palacio et al., 2016; Leslie et al., 2005a), F. pseudonygamai (Leslie et al., 2005a), and F. semitectum (Prom et al., 2015) although their role in pathogenicity is not as clearly defined as it is for the species described above. None of these species were recovered from our samples of Thai sorghum. Non-cultivated Sorghum species can be colonized by an even larger number of Fusarium species, e.g., Walsh et al. (2010).
Fusarium thapsinum is known to be an important cause of stalk rot and grain mold of sorghum (Jardine and Leslie, 1992; Klittich et al., 1997; Leslie et al., 2005a; Little et al., 2012; Onyike and Nelson, 1992). Although it can be recovered from numerous other plants, e.g., native grasses (Bentley et al., 2007; Leslie et al., 2004), date palms (Al-Sadi et al., 2012), maize (Klittich et al., 1997; Leyva-Madrigal et al., 2015; Madania et al., 2013; Sampietro et al., 2010), wheat (Busman et al., 2012), and soybeans (Pedrozo and Little, 2014). Sorghum, though, appears to be a preferred host for F. thapsinum, and the fungus has been previously reported from sorghum in Africa (Klittich et al., 1997), the Philippines (Klittich et al., 1997), India (Sharma et al., 2011), Korea (Choi et al., 2013), Australia (Petrovic et al., 2009), and the Americas (Klittich et al., 1997; Sampietro et al., 2010).
The relative frequencies of F. proliferatum, F. thapsinum, and F. verticillioides in this study were approximately the same, in contrast to the frequencies of these species recovered from sorghum in Kansas, where F. thapsinum is the most frequent, F. proliferatum is next, and isolates of F. verticillioides are relatively rare. In Thailand, sorghum usually is planted as a second crop after maize or rice is harvested (Salleh et al., 1995). Thus, the increased frequencies of F. proliferatum and F. verticillioides could be due to inoculum carryover from a previous crop. F. proliferatum is common on both rice and maize, and F. verticillioides is common on maize, where these fungi may be hosted either as endophytes or as disease-causing agents (Cartwright et al., 2018; Leslie and Summerell, 2006; Munkvold and White, 2016). In sorghum, both F. verticillioides and F. proliferatum can cause stalk and root rot (Jardine and Leslie, 1992; Palmero et al., 2012), and F. proliferatum also can cause grain mold (Sharma et al., 2011; Tesfaendrias et al., 2011). The broad host range of F. proliferatum enables this fungus to survive not only on rice, maize, mangoes and sorghum (Leslie, 1995; Mohamed Nor et al., 2013), but on many other plants grown in Thailand and provides numerous potential and widely dispersed inoculum sources. Fusarium verticillioides and F. thapsinum are both likely introductions to Thailand, with F. verticillioides arriving with maize and F. thapsinum with sorghum. F. proliferatum, a known pathogen of rice, probably originated in this general geographic region and has spread to other crops as Thai agriculture has diversified.
We also recovered a few strains of F. sacchari and F. beomiforme in this study. F. sacchari is not a common problem on sorghum. This species is best known as the cause of a sorghum leaf disease (Leslie et al., 2005b; Sharma et al., 2011), although it has been associated with sorghum crown and root rot in Australia (Petrovic et al., 2013). Fusarium beomiforme is the only species we recovered that does not belong to the Fusarium fujikuroi species complex, from which it is excluded phylogenetically (Laurence et al., 2011). This fungus has been recovered from soil and soil debris in Australia, South Africa, and Papua New Guinea (Nelson et al., 1987). In Papua New Guinea, F. beomiforme was recovered from soil in which sorghum had been cultivated (Nelson et al., 1987), but to date, F. beomiforme has not been reported to colonize or to be pathogenic to sorghum. Our recovery of F. beomiforme from sorghum tissue in this study expands the range of plants from which this species has been recovered.
One important species associated with sorghum pathogenicity, Fusarium andiyazi (Leslie et al., 2005a; Marasas et al., 2001), was not identified in our survey although it has been reported from rice growing in Malaysia (Hsuan et al., 2011). This species has been isolated from sorghum in South Africa, Ethiopia, Nigeria, and the USA (Marasas et al., 2001). The absence of F. andiyazi could indicate that much of the sorghum in Thailand has a recent external origin, perhaps from India, where this species has not been reported, rather than from Africa, where it is widespread. Relatively little is known concerning the host range and pathogenicity of F. andiyazi, so the risk to Southeast Asian agriculture remains difficult to assess. Monitoring the presence of any imported African sorghum for the presence of this species should be considered as should more comprehensive surveys of sorghum and rice for this pathogen, especially in areas outside of peninsular Thailand.
Isolates in the same Fusarium species generally share 65% or more of their AFLP bands. Based on the AFLP bands, two strains of F. verticillioides, two of F. thapsinum, and four of F. proliferatum, appear to be clones, i.e., the strains in each clonal group have the same AFLP banding pattern. Members of a clonal group were all collected at the same location and belonged to the same VCG (Table 2).
Effective population number (Ne) and AMOVA analyses both provide information on the populations of F. proliferatum, F. thapsinum, and F. verticillioides. The population samples are small for all of the species identified, however, and must be analyzed with some caution. Ne based on mating type is usually larger than Ne based on female fertility, and commonly exceeds 90% of the count (Leslie and Klein, 1996). In this study, Ne(mt) was larger than Ne(f) for all three species (Table 3) and exceeded 90% of the count for both F. proliferatum and F. verticillioides. The Ne(mt) value of 75% of the count for F. thapsinum is lower than usually observed for this species. Ne(f) was high, 98% of the count, for F. verticillioides, and much lower, between 40 and 50% of the count, for F. proliferatum and F. thapsinum. These results suggest that sexual reproduction is an important part of the life cycle of F. verticillioides in Thailand, as maintaining high levels of female fertility is disadvantageous, at least in the short term, if a more efficient asexual means of reproduction is available. The level of female fertility in F. proliferatum usually is lower than in F. verticillioides. The values observed in this study suggest that this species reproduces extensively via asexual reproduction in Thailand, as the proportion of the population participating in sexual reproduction is probably 1–2% at any given time. This number is significantly less than previously reported for a global population of F. proliferatum (Leslie and Klein, 1996). Sexual reproduction is not of primary importance for F. thapsinum at any location, as assessed by Ne(f), since the number of female fertile strains always is low. Ne(f) for this Thai population, 42% of the count, which is somewhat higher than the 32% previously reported (Leslie and Klein, 1996), and strengthen the current view of F. thapsinum as a species that maintains just enough sexual reproduction to avoid losing sexual reproductive capacity altogether.
The AMOVA analysis (Table 4) also presents different pictures for each of the three main species. The F. thapsinum populations are the best connected as 83% of the variation is found “within populations” and only 17% is found “among populations”. This result could occur if F. thapsinum is limited to sorghum in Thailand and inoculum is distributed with seed. The bottleneck effect that comes from subdividing a larger, relatively uniform, population would then result in the relatively small observed “among population” variation. There are no other published reports of F. thapsinum occurring in Thailand. This void combined with the observed distribution of population variation suggests that F. thapsinum is not a particularly effective competitor of other grain crops grown in Thailand, e.g. rice and maize, where more extensive studies of Fusarium are available, e.g., Darnetty and Salleh (2017). However, the common identification by Thai researchers of many strains of Fusarium as F. moniliforme, even when molecular techniques are used, could be hiding the presence of F. thapsinum, since F. thapsinum and F. verticillioides, are very similar morphologically (Leslie and Summerell, 2006) and at one time were both called F. moniliforme (Klittich and Leslie, 1992; Klittich et al., 1997; Seifert et al., 2003). Similarly, strains identified as F. oxysporum that cause disease on sorghum, e.g., Kavitha et al. (2010) might also be strains of F. thapsinum.
Fusarium proliferatum populations were much less connected as nearly a third of the population variance was “among populations”. This result was not expected since this species is endemic to Thailand rather than introduced. It could result from F. proliferatum populations originating from local sources and perhaps from multiple non-sorghum hosts.
The F. verticillioides populations were the most isolated of all, with 47% of the variance “among populations” and only 53% of the variance “within populations”. Thus, more than half of the total genetic variance observed is not shared between different populations. Although possible, it is unlikely that this introduced species was distributed with the sorghum seed, as this species does not usually colonize sorghum as an endophyte or preferred host (Leslie and Summerell, 2006). Sorghum planted into fields where an earlier crop was maize could be infected by F. verticillioides isolates that had persisted in the soil, as F. verticillioides is a very common endophyte and pathogen of maize in Thailand (Anukul et al., 2014; Darnetty and Salleh, 2017). There are numerous sources of commercial maize in Thailand, and different suppliers could distribute (unintentionally) different populations of F. verticillioides with their seed. As these seed producers would have distinct operations, the F. verticillioides populations distributed with the seed probably would not have large overlaps. The distinctness of the sampled fungal populations could result from farmers consistently purchasing seed from the same commercial supplier or carrying seed over from year-to-year, and consistently (re-)introducing the same or very similar fungal populations. Alternatively, this pattern could result if the ability of F. verticillioides to persist in the local soil is limited, and only the most recently introduced population was present to colonize the succeeding crop, in this case sorghum.
The key difference between the sampled F. thapsinum and F. verticillioides populations is that the F. verticillioides population must have been present in the field before sorghum was planted there, as F. verticillioides is not a common endophyte of sorghum, and may have been there for some time as a pathogen/endophyte/saprophyte of other suitable hosts. The uniformity of variation within the F. thapsinum populations suggests that all of the sampled populations arose relatively recently from a common source and that most, if not all, of the members of these populations have been distributed from that common source to the sampled fields.
Mycotoxin contamination of grain is not usually a major problem for sorghum, although it can occur (Leslie, 2014). The three commonly recovered species in this survey, i.e., F. proliferatum, F. verticillioides, and F. thapsinum, all can produce fumonisins, although the amounts produced by F. thapsinum usually are quite small (Desjardins, 2006; Leslie and Summerell, 2006; Leslie et al., 2005a). Sorghum contaminated with fumonisins can be hazardous to humans and domesticated animals. These species also can produce another mycotoxin, moniliformin. F. thapsinum is the most prolific moniliformin producer of these three species (Leslie et al., 2005a). Moniliformin is toxic to chickens, and has been associated with Keshan heart disease in China (Desjardins, 2006). Thus, sorghum produced in Thailand could potentially be contaminated with either fumonisins or moniliformin, and those using this grain for animal feed should be aware of the possibility of contamination by these mycotoxins and the symptoms of toxicosis that may occur in animals consuming contaminated feed(s).
In summary, we have recovered four Fusarium spp. within the F. fujikuroi species complex and one additional species from sorghum fields in Thailand. Fusarium thapsinum, F. proliferatum, F. sacchari, and F. verticillioides are from the F. fujikuroi species complex and have been previously associated with sorghum. Three of these species (F. thapsinum, F. proliferatum, and F. verticillioides) are potential toxigenic species, and should be managed to reduce the amount of toxin-contaminated grain in the field. The structure of the populations for the three most common species suggests that these species are not transmitted to the plant in the same manner. Reduction in contamination by F. thapsinum, for example, might be implemented relatively simply through more stringent plant health efforts with seed grown locally or imported for the next season’s crops. Reductions in contamination by F. proliferatum or F. verticillioides, however, might be more difficult to achieve as they are well-established on other crops grown widely within the country. Confirming the channels through which these species are being transmitted would increase our understanding of the larger agricultural systems in Southeast Asia, and of cultural practices that could alter the local populations of these pathogens on sorghum and other cultivated crops.
Acknowledgments
We thank Wiwut Daengsubha for assistance collecting samples, and Amy Beyer and Bruce Ramundo for technical laboratory assistance. This research was supported in part by the Malaysian Ministry of Higher Education through the Academic Staff Training Scheme of the Universiti Sains Malaysia and Fundamental Research Grant Scheme (Project number: 203/PBIOLOGI/6711576), INTSORMIL – the International Sorghum and Millets Collaborative Research Support Program (AID/DAN-1254-G-00-0021-00), and USDA National Institute of Food and Agriculture Hatch Multi-state project KS1183A. Manuscript no. 19-301-J from the Kansas Agricultural Experiment Station, Manhattan.