Optimization of Polyethylene Glycol-Mediated Transformation of the Pepper Anthracnose Pathogen Colletotrichum scovillei to Develop an Applied Genomics Approach
Article information
Abstract
Colletotrichum acutatum is a species complex responsible for anthracnose disease in a wide range of host plants. Strain C. acutatum KC05, which was previously isolated from an infected pepper in Gangwon Province of South Korea, was reidentified as C. scovillei using combined sequence analyses of multiple genes. As a prerequisite for understanding the pathogenic development of the pepper anthracnose pathogen, we optimized the transformation system of C. scovillei KC05. Protoplast generation from young hyphae of KC05 was optimal in an enzymatic digestion using a combined treatment of 2% lysing enzyme and 0.8% driselase in 1 M NH4Cl for 3 h incubation. Prolonged incubation for more than 3 h decreased protoplast yields. Protoplast growth of KC05 was completely inhibited for 4 days on regeneration media containing 200 μg/ml hygromycin B, indicating the viability of this antibiotic as a selection marker. To evaluate transformation efficiency, we tested polyethylene glycol-mediated protoplast transformation of KC05 using 19 different loci found throughout 10 (of 27) scaffolds, covering approximately 84.1% of the entire genome. PCR screening showed that the average transformation efficiency was about 17.1% per 100 colonies. Southern blot analyses revealed that at least one transformant per locus had single copy integration of PCR-screened positive transformants. Our results provide valuable information for a functional genomics approach to the pepper anthracnose pathogen C. scovillei.
Pepper (Capsicum annuum L.) belongs to the genus Capsicum in the Solanaceae family and is considered an economically important and popular vegetable crop. There are five domesticated species of peppers: C. annuum L., C. frutescens L., C. chinense Jacq., C. pubescens Ruiz & Pav., and C. baccatum L. (Kraft et al., 2014). Among these, C. annuum is the most common and widely cultivated species worldwide (Oo et al., 2017, Saxena et al., 2016). In Korea, pepper is one of the most popular vegetables; approximately 72 thousand tons of peppers were produced in Korea in 2018, an increase of 16 thousand tons (28.4%) from 56 thousand tons in 2017 (KOSTAT, Statistics Korea, http://kostat.go.kr/portal/eng/). Pepper is valued for its beneficial effects on human wellness; it contains organic micronutrients, including carotenoids, flavonoids, and vitamins A, C, and K, and this vegetable helps to reduce blood pressure and cholesterol levels (Dias, 2012). In addition to its health benefits, pepper is used fresh, as a spice, or as a minor ingredient in various dishes such as hot soup and kimchi.
Many fungal pathogens are known to cause diseases in pepper, including Colletotrichum species, which cause anthracnose disease; Rhizoctonia solani, which causes rhizoctonia root rot; and Phytophthora capsici and P. nicotianae, which cause phytophthora blight (Chi et al., 2013; Mannai et al., 2018; Than et al., 2008). Among these diseases, anthracnose is the most devastating fungal disease of pepper worldwide; yield losses of pepper are estimated at more than $100 million USD in Korea (Oo et al., 2017; Than et al., 2008). The genus Colletotrichum contains approximately 189 species. Of these, 34 species are recognized within the C. acutatum species complex (Baroncelli et al., 2017). Many Colletotrichum species, including C. acutatum, C. gloeosporioides, and C. capsici, infect pepper (Bailey and Jeger, 1992; Than et al., 2008). Colletotrichum acutatum is one of the most frequently reported species causing anthracnose disease on pepper in tropical and subtropical countries (Han et al., 2016). Using combined sequence analyses of the β-tublin-2 (TUB2) gene, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, and internal transcribed spacer (ITS) rDNA regions, C. scovillei (a member of the C. acutatum species complex) was recently shown to cause anthracnose disease on pepper in several countries including Brazil, China, Japan, and South Korea (Caires et al., 2014; Diao et al., 2017; Kanto et al., 2014; Zhao et al., 2016).
Fungal transformation can be a powerful tool in functional genomics (Fincham, 1989; Hynes, 1996). This technique enables researchers to understand the complex mechanisms of fungal infection and fungal pathogenesis at the molecular level. Most fungal transformation methods include the formation of fungal protoplasts that have had their cell walls removed using mechanical or enzymatic methods (Hayat and Christias, 2010; Song et al., 2015). Several factors influence the efficiency of protoplast generation, including fungal strain, culture media, culture temperature, culture pH, age of mycelia, enzyme concentrations, enzyme types, and osmatic stabilizers (Chadegani et al., 1989; Eyini et al., 2006). Cellulase, 1,3-glucanase, chitinase, Trichoderma harzianum lysing enzyme, and driselase are frequently used as cell wall degrading enzymes (Hayat and Christias, 2010; Song et al., 2015; Xiao et al., 2013).
The factors affecting protoplast regeneration and transformation efficiency include type of antibiotics, vector size, form of DNA (PCR-amplified DNA, pre-linearized plasmid, or circular form plasmid), concentration of DNA, and method of transformation. Genetic transformation can be conducted using different transformation methods, including electroporation, Agrobacterium-mediated transformation, polyethylene glycol (PEG)-mediated transformation, and the Cas9 ribonucleoprotein gene editing system (Chung et al., 2002; Liu and Friesen, 2012; Moradi et al., 2013; Wang et al., 2018b). Among these, PEG-mediated transformation is a particularly simple and efficient method. In this study, we developed a transformation system of C. scovillei KC05 by optimizing the factors affecting protoplast generation, transformation, and regeneration. Our results will provide valuable tools for research of the molecular mechanisms of the pepper pathogen C. scovillei KC05.
Materials and Methods
Fungal strain identification
Colletotrichum acutatum KC05 isolated from an infected pepper fruit in Gangwon Province of South Korea was used in this study (Han et al., 2016). The isolate was identified based on combined sequence analyses of the nuclear ribosomal internal transcribed (ITS) region, β-tubulin (TUB2), actin (ACT), partial sequences of the chitin synthase 1 (CHS-1), an intron sequence of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and histone 3 (HIS3) genes (Damm et al., 2012; da Silva et al., 2017). The six gene sequences of C. acutatum KC05 were obtained from Han et al. (2016), and the gene sequences of other Colletotrichum species (C. acutatum, C. scovillei, and C. simmondsii) were obtained from GenBank (Supplementary Table 1). Percentage sequence identities of the six genes were obtained from the National Center for Biotechnology Information database (NCBI) using nucleotide BLAST. A phylogenetic tree was generated using maximum likelihood analyses based on combined analyses of the six gene sequences of the selected Colletotrichum species.
Colletotrichum scovillei KC05 protoplast generation
Fungal conidia for protoplast generation were collected from a 10-day-old oatmeal medium agar (5% oatmeal and 2.5% agar) culture, inoculated into 100 ml complete medium broth (0.6% yeast extract, 0.6% casamino acids, and 1% sucrose), and cultured on a shaker at 150 rpm for 1 day at 25°C in the dark. The resultant mycelia were harvested onto a sterilized cheese cloth and washed twice with 20% sucrose. Lysing enzyme and driselase were suspended in 1 M NH4Cl and sterilized through 20 μm pore filters (Sigma, St. Louis, MO, USA). To evaluate the enzymatic effects for protoplast generation, the washed mycelia were divided into 1,500 mg aliquots in 50 ml conical centrifuge tubes (SPL, Pocheon, Korea) and resuspended with 10 ml 1 M NH4Cl containing different lytic enzymes. Then the suspension was incubated at 25°C with gentle shaking at 70 rpm, and protoplasts were counted using a hemocytometer. Protoplasts were pelleted by centrifugation at 5,000 rpm, washed, and suspended individually in 1.5 ml microcentrifuge tubes containing 200 μl 1× STC buffer (40% sucrose, 1 M Tris-HCl, and 1 M CaCl2). A differential interference contrast microscope (Axio Imager.A2, Zeiss, Jena, Germany) was used to examine the shapes of protoplasts. Protoplasts were stored at −75°C. Experiments were conducted in triplicate and repeated three times. All data were processed using the SigmaStar statistical software package (SPSS Science, Chicago, IL, USA), and error bars represent 95% confidence intervals.
Sensitivity of C. scovillei KC05 protoplasts to antibiotics
Hygromycin B (Millipore, Billerica, MA, USA) and geneticin (Gibco Life Technologies, Carlsbad, CA, USA) were used as selection antibiotics. Aliquots (200 μl) of protoplast suspension were transferred to 15 ml conical centrifuge tubes, mixed with an equal volume of PTC buffer (2× STC plus 40% PEG), incubated for 20 min at room temperature, and gently shaken in a rotary shaker for 5 h. Different concentrations of antibiotics and incubated protoplast aliquots were added to regeneration media (20% sucrose, 0.3% yeast extract, 0.3% casamino acids, 1% glucose, and 0.8% agar; autoclaved and cooled to 55°C) and poured into Petri dishes (90 mm in diameter). Plates were incubated at 25°C in the dark for 4 days.
Protoplast transformation
Nineteen loci were selected from 10 of 27 different scaffolds (Han et al., 2016). For double-joint PCR and Southern blots, genomic DNA was extracted using a quick DNA extraction method (Chi et al., 2009). Gene knock-out constructs were generated using double-joint PCR (Park et al., 2014). Primer sequences used in this study are listed in Supplementary Table 2. The hygromycin phosphotransferase gene (HPH) cassette was amplified using the primers HPH_F (5′-GGCTTGGCTGGAGCTAGTGGAGG-3′) and HPH_R (5′-CTCCGGAGCTGACATCGACACCAAC-3′) from pBCATPH (Kim et al., 2009), and approximately 1.4 kb of two flanking regions of target loci were amplified using the primers 5F/R and 3F/R. The two fragments of flanking regions and the HPH cassette were fused using double-joint PCR, and the final construct was amplified using the primers NF and NR. Five micrograms of the construct haboring hygromycin resistance gene cassettes were introduced into 200 μl C. scovillei KC05 protoplasts (1 × 107 protoplasts/ml) using PEG-mediated transformation (Park et al., 2014; Sweigard et al., 1992). Transformed protoplasts and hygromycin B (200 μg/ml) were mixed with 100-ml regeneration media (autoclaved and cooled to 55°C) and poured into 90-mm Petri dishes. The number of colonies growing on the regeneration media was counted 4 days after transformation, and PCR screening was performed using the primers SF/SR. Southern blot was performed according to standard procedures to determine the number of copies of the transformed construct (Shin et al., 2014).
Results
Fungal strain identification
Colletotrichum acutatum is a species complex with many members (Baroncelli et al., 2017). The previously identified strain C. acutatum KC05 (Han et al., 2016) was used in the present study to confirm the identity in C. acutatum sensu lato. Gene sequences of ITS, TUB2, ACT, CHS-1, GAPDH, and HIS3, which are mainly used in species classification (Caires et al., 2014), were analyzed in C. acutatum KC05, for comparison with those in C. scovillei CBS 126529, C. simmondsii CBS 122122, C. fioriniae CBS 128517, C. acutatum CBS 112996, C. graminicola CBS 112999, C. higginsianum IMI 349063, C. gloeosporioides CBS 112999, and C. orbiculare CBS 104-T. This analysis revealed that the ITS, TUB2, ACT, GAPDH, and HIS3 gene sequences of C. scovillei KC05 had 100% identity matches with C. scovillei CBS 126529, and the CHS-1 sequence exhibited 99% identity matches (Table 1). In the five multigene analyses, C. scovillei KC05 showed a higher match (> 90% identity) with members in C. acutatum sensu lato, C. scovillei, C. simmondsii, C. fioriniae, and C. acutatum, compared to the other species, C. graminicola, C. higginsianum, C. gloeosporioides, and C. orbiculare. A phylogenetic tree was subsequently constructed using the multigene sequences to analyze the genetic relationship among species.
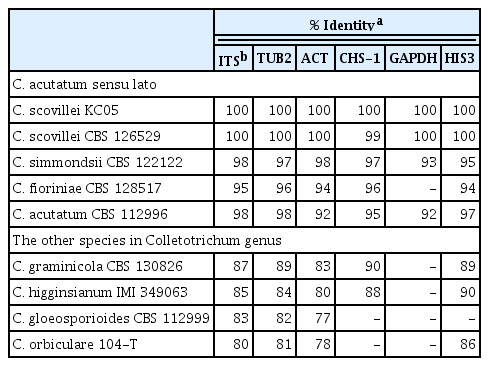
Percentage sequence identities of ITS, TUB3, ACT, CHS-1, GAPDH, and HIS3 genes from Colletotrichum species
As expected, C. scovillei KC05 was grouped in a clade with the members of C. acutatum sensu lato, C. scovillei, C. simmondsii, C. fioriniae, and C. acutatum, in which C. scovillei KC05 was almost identical to the reference strain C. scovillei CBS 126529 (Fig. 1). This result is consistent with that of Oo et al. (2017) and further indicates that C. acutatum KC05 is C. scovillei.

Combined internal transcribed spacer (ITS), β-tubulin (TUB2), actin (ACT), chitin synthesis 1 (CHS-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and histone 3 (HIS3) gene sequences. The phylogenetic tree illustrates relationships among Colletotrichum species in C. scovillei KC05 isolated from Korean pepper. Numbers at nodes represent the percentages of occurrence in 1,000 bootstrap replicates. Scale bar indicates the number of nucleotide substitutions.
Colletotrichum scovillei KC05 protoplast generation
As a step toward understanding pepper anthracnose disease, we optimized a transformation system of C. scovillei KC05. For protoplast generation of C. scovillei KC05, two commonly available enzymes, lysing enzyme and driselase, were tested on C. scovillei KC05 mycelia. At 1 h after incubation of C. scovillei KC05 mycelia with lysing enzyme, 6.0 × 104 protoplasts/ml were released with 0.5% lysing enzyme (Fig. 2). The addition of more enzyme enhanced protoplast yields. Averages of 13.0 × 104 and 17.6 × 104 protoplasts/ml were released from C. scovillei KC05 mycelia with 1% and 2% lysing enzyme, respectively. Longer incubation times also enhanced protoplast yields. At 3 h after incubation with 0.5% lysing enzyme, 75 × 104 protoplasts/ml were released, which was 12.5-fold higher than after 1 h incubation. However, prolonged incubation for more than 3 h resulted in protoplast cell lysis; only 33.3 × 104 protoplasts/ml were released from mycelia at 5 h after incubation, which was approximately 2.2-fold lower than at 3 h incubation. Driselase released lower numbers of protoplasts compared to lysing enzyme. The maximum release of protoplasts with driselase was obtained with 0.8% driselase at 3 h incubation, which released approximately 58 × 104 protoplasts/ml. A combination of the two enzymes released more protoplasts than just one enzyme alone; 2% lysing enzyme and 0.8% driselase with 3 h incubation produced the maximum release of protoplasts, 860 × 104 protoplasts/ml.
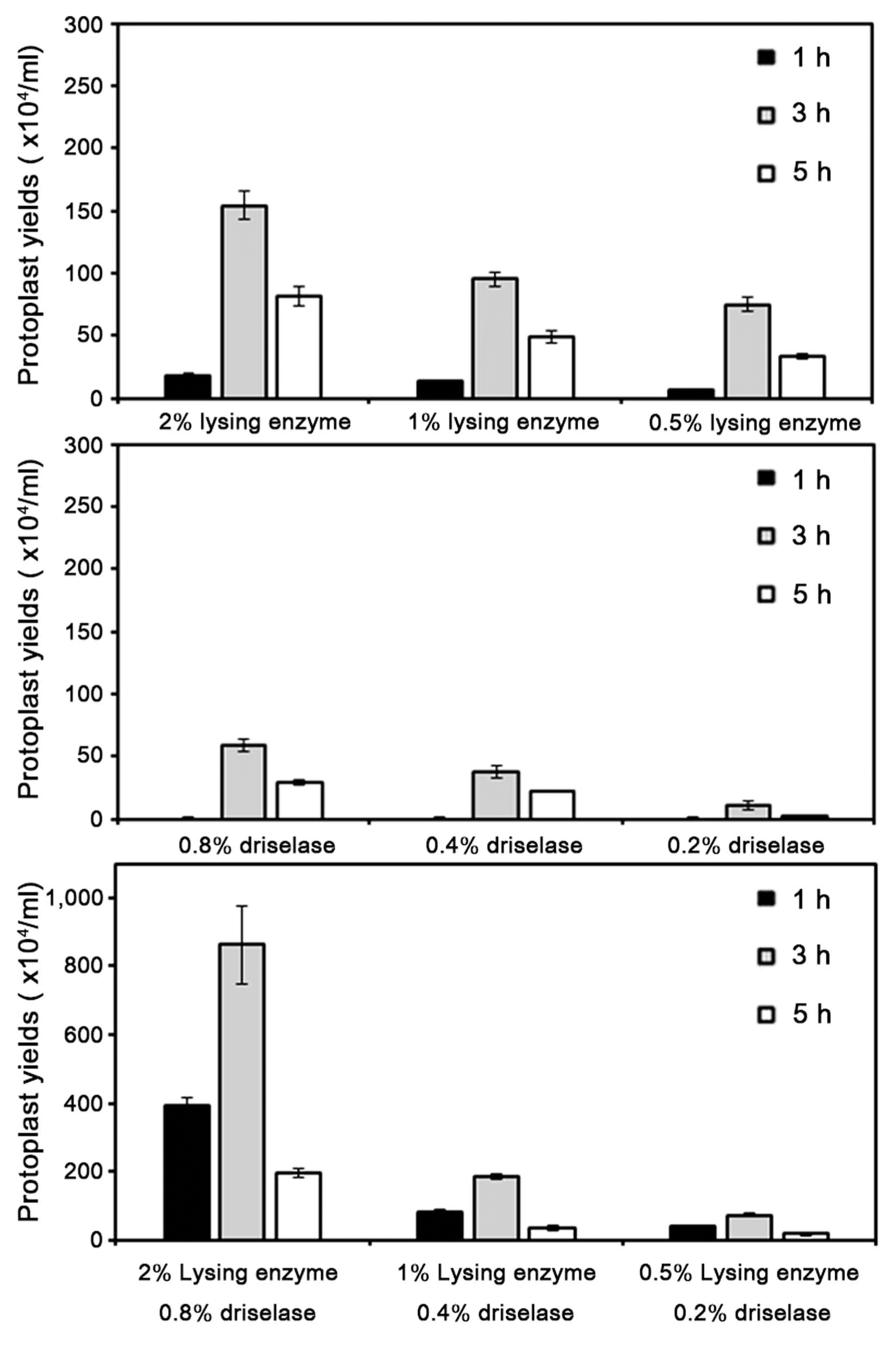
Effect of different concentrations of lysing enzyme or driselase on protoplast yields of Colletotrichum scovillei KC05. Washed mycelia were divided into 1,500 mg aliquots in 50 ml conical centrifuge tubes, resuspended with 10 ml 1 M NH4Cl containing different lytic enzymes, and incubated at 25°C with gentle shaking at 70 rpm. Experiments were conducted in triplicate and repeated three times. Error bars indicate standard deviations.
The shapes of released protoplasts were spherical, and their average size was 8.82 ± 0.06 μm in diameter after 3 h incubation in the 2% lysing enzyme and 0.8% driselase mixture (Fig. 3). After 5 h incubation in the same mixture, the average size of protoplasts was 11.09 ± 0.07 μm, much larger than at 3 h incubation. Noticeably, lysed protoplasts were observed after 5 h incubation. These results indicate that the combination of lysing enzyme and driselase at 3 h incubation is most effective for protoplast generation from mycelia of C. scovillei.
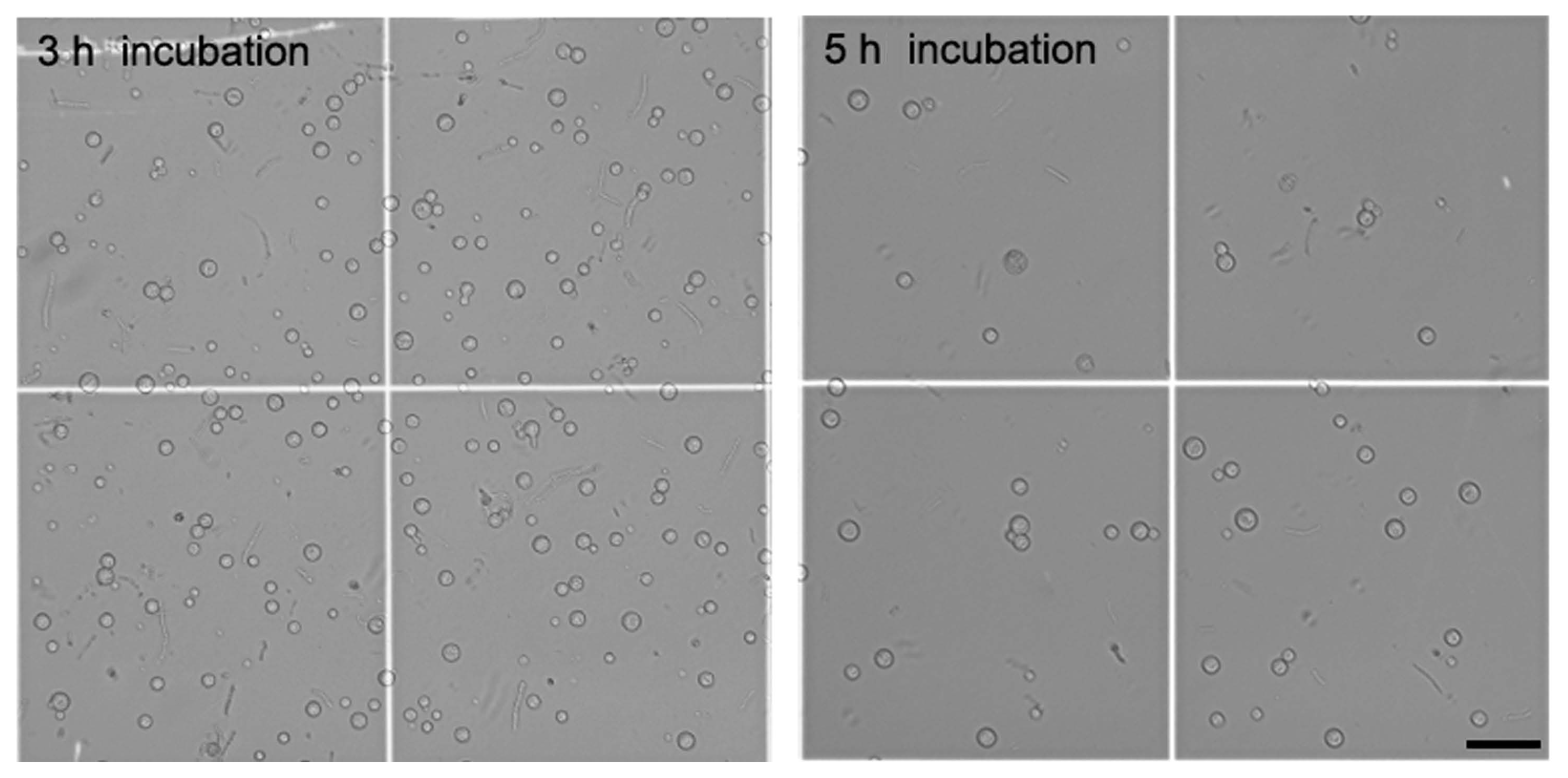
The shape of released protoplasts of Colletotrichum scovillei KC05. The average size of protoplasts was 8.82 ± 0.06 μm after 3 h incubation and 11.09 ± 0.07 μm after 5 h incubation in the 2% lysing enzyme and 0.8% driselase mixture. Lysed protoplasts were observed after 5 h incubation under a differential interference contrast microscope. Scale bar = 50 μm.
Sensitivity of C. scovillei KC05 protoplasts to antibiotics
The sensitivity of C. scovillei KC05 protoplasts to antibiotics was assessed to select a positive transformation marker and to determine the minimal concentration of antibiotics required. Hygromycin B and geneticin were used as the selective antibiotics, as both have been widely used for fungal transformation. Results indicated that hygromycin B completely inhibited the growth of C. scovillei KC05 protoplasts at a concentration of 200 μg/ml (Fig. 4). However, at lower concentrations (50 and 100 μg/ml), protoplast growth was not completely inhibited. Compared to hygromycin B, protoplasts were more resistant to geneticin; protoplast growth was not completely inhibited at concentrations of either 200 or 400 μg/ml. However, 800 μg/ml geneticin was able to completely inhibit protoplast growth. Based on these results, different concentrations of the two antibiotics were used in the selection of transformants of C. scovillei KC05.

Sensitivity of Colletotrichum scovillei KC05 protoplasts to antibiotics. Hygromycin B and geneticin were used as selection antibiotics. Different concentrations of antibiotics and protoplasts were added to regeneration media and poured into 90-mm Petri dishes. Plates were incubated at 25°C in the dark for 4 days.
Protoplast transformation
To better understand the transformation efficiency of C. scovillei KC05, a total of 19 loci were randomly selected from the KC05 genome (Han et al., 2016). These loci were scattered throughout 10 (out of 27) different scaffolds, which covered 49.9 Mb, equivalent to 84.1% of the whole genome (Fig. 5). For the homology-dependent replacement of a targeted gene, knock-out DNA constructs were generated for the 19 loci using PCR. Protoplasts of KC05 were transformed with DNA of each knock-out construct (Figs. 5 and 6A). Transformed protoplasts were grown on regenaration media in 90-mm Petri dishes supplemented with hygromycin B. Many transformants were regenerated at 4 days after transformation, and 45–138 transformants were subsequently selected to simply screen putative knock-out transformants using a PCR method (Table 2). The PCR screening was performed with the primers SF/SR using genomic DNA extracted from the transformants (Fig. 6B). Two bands at 1.7 and 3.9 kb were amplified within many transformants (Fig. 6B), as illustrated in Fig. 6A. These results suggest that the targeted gene remained intact and that the DNA constructs were ectopically integrated in the genome. However, the detection of only the 1.7 kb band in several transformants suggests possible knock-out candidates via a homology-dependent replacement event. By measuring one band in transformants for the 19 loci, we obtained a variable number of positive candidates, ranging from 5 to 26 (Table 2). For example, 24 of 81 transformants were positively detected as knock-out candidates for the CSP_010985 locus, while 8 of 111 transformants were positive in the PCR screening for CSP_006969. The average number of candidates for knock-out mutants was 17.1% for the 19 loci tested in the PCR screening. To confirm the target gene-specific replacement in knock-out transformants, Southern blot analyses were performed with transformants. As illustrated in Fig. 6A, genomic DNA of transformants was digested using NdeI and then hybridized with the indicated probe. The analyses revealed that 4.0 kb NdeI bands were only detected in knock-out mutants, while 6.7 and 2 kb bands originated from transformants with ectopic integration and the wild-type copy of the gene, respectively (Fig. 6C). Knock-out mutants were obtained for different sizes of the 19 targeted loci (Table 2).
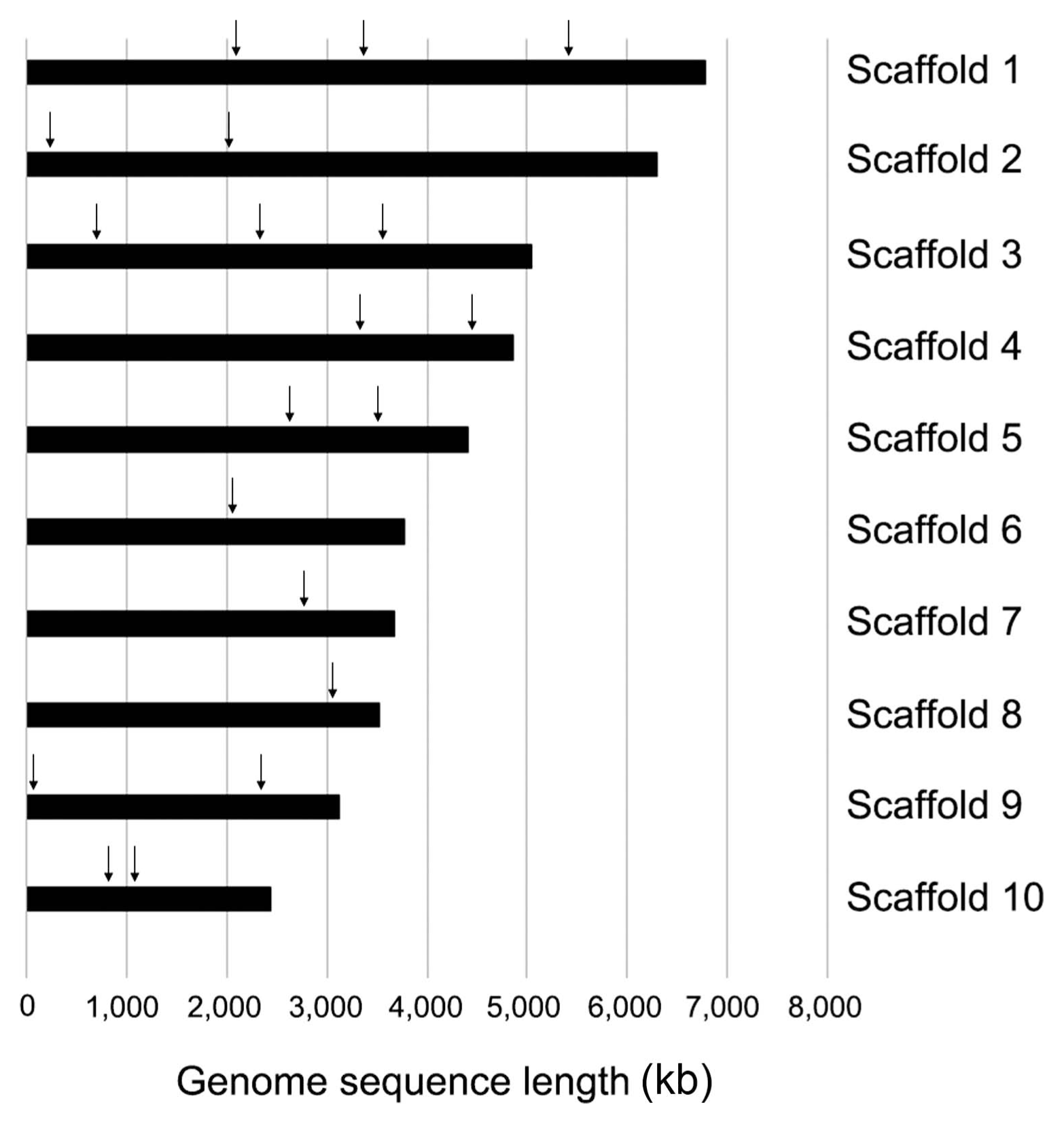
Distribution of 19 loci in 10 scaffolds of Colletotrichum scovillei KC05. Scaffold 1 (annotation numbers: CSP_000583, CSP_000836, and CSP_001376; scaffold 2 (CSP_001741 and CSP_002212); scaffold 3 (CSP_003491, CSP_003850, and CSP_004175); scaffold 4 (CSP_005482 and CSP_005764); scaffold 5 (CSP_006607 and CSP_006807); scaffold 6 (CSP_007586); scaffold 7 (CSP_008738); scaffold 8 (CSP_009802); scaffold 9 (CSP_010617 and CSP_009947); and scaffold 10 (CSP_010985 and CSP_011033). Each locus is ordered from left to right in each scaffold bar.

Construction of a gene knock-out system and identification of deletion mutant. (A) CSP_006607 gene (gray arrow) was deleted through the targeted gene replacement method. (B) PCR screening was performed using the primers SF/SR. The primer pair produced a 1.7-kb band from gene knock-out transformants (indicated by an asterisk) and both 1.7- and 3.9-kb bands from random insertional transformants. (C) Southern blot analyses. DNA samples were digested with NdeI. A 6.7-kb band was produced from wild type KC05, and a 4.0-kb band was produced from a knock-out mutant. Lane 1, wild type; lane 2, ectopic; lane 3, knock-out mutant; and lane 4, ectopic.
Discussion
C. acutatum is an important anthracnose pathogen commonly identified from a wide range of host plants. Several studies have recently identified subgroups within the C. acutatum species complex, using the combined gene sequences of ITS, TUB2, ACT, CHS-1, GAPDH, and HIS3 (Bragança et al., 2016; Damm et al., 2012). For example, C. scovillei is proven to be separate species of the C. acutatum species complex through multigene sequence analysis (Damm et al., 2012). Within the C. acutatum species complex, C. scovillei has frequently been identified as a causal agent, most frequently from fruit-rots including peppers. Supportively, Oo et al. (2017) collected 35 isolates of Colletotrichum sp. from infected pepper fruits in South Korea, and subsequently identified the isolates using the TUB2, GAPDH, and ITS sequences of strain CBS 126529. All of these isolates were identified as C. scovillei. In the present study, C. acutatum KC05 isolated from infected pepper fruits in South Korea was idenfied as C. scovillei using the ITS, TUB2, ACT, CHS-1, GAPDH, and HIS3 sequences. Based on our results and those of previous studies, we concluded that C. scovillei is an important anthracnose pathogen of pepper fruits in South Korea. In a previous study, we collected 30 Colletotrichum strains from pepper and apple fruits in several fields of Gangwon Province (data not shown). Through analyses of ITS sequences, we confirmed that the isolates were C. gloeosporioides, C. acutatum, C. graminicola, and C. fioriniae. In future studies, we plan to collect more Colletotrichum species from various fruits to then identify isolates using combined sequence analyses of the ITS, TUB2, ACT, CHS-1, GAPDH, and HIS3 genes.
The key factors affecting protoplast release of filamentous fungi include the type of enzymes, enzyme combinations, age of mycelia, and incubation time (da Silva Coelho et al., 2010; Xiao et al., 2013). Rehman et al. (2016) optimized the protoplast isolation, regeneration, and transformation efficiency of Verticillium dahlia, the causal agent of Verticillium wilt. The maximum release of protoplasts was obtained using 200 mg driselase in 10 ml NaCl (0.7 M) at 2.5 h after incubation. Similar to our data, prolonged incubation times resulted in lysing of protoplasts. Cheng and Bélanger (2000) described a protocol for yielding protoplasts from Pseudozyma flocculosa, a powdery mildew biocontrol agent. In their experiment, 0.5% Novozyme 234 was the most efficient enzyme, followed by 5% Glucanex. The maximum release of protoplasts was obtained from a combination of 0.5% Novozyme 234 and 5% Glucanex (Cheng and Bélanger, 2000). Ramamoorthy et al. (2015) evaluated various cell wall degrading enzymes for the production of protoplasts in Fusarium verticillioides and concluded that a combination of lysing enzyme and driselase was effective for protoplast production. In the present study, we determined that the combination of 2% lysing enzyme and 0.8% driselase in 1 M NH4Cl with 3 h incubation was optimal for protoplast release in C. scovillei KC05. Consistent with these previous studies, prolonged incubation times resulted in the generation of fewer protoplasts, and the combination of different enzymes resulted in the release of more protoplasts than with one enzyme alone.
Prior to transformation, a suitable selection marker must be chosen for transformation. Hygromycin B is the most commonly used antibiotic as a selection marker for tranformation in Ascomycota (Yörük and Albayrak, 2015). Geneticin, on the other hand, is used for complement transformation (Han et al., 2015, Wang et al., 2018a). In the present study, we selected hygromycin B and geneticin as selective markers and evaluated their effectiveness for the growth inhibition of C. scovillei KC05 protoplasts. Chung et al. (2013) generated gene deletion mutants in Magnaporthe oryzae via selection on regeneration media supplemented with hygromycin B (200 μg/ml concentration) or geneticin (800 μg/ml concentration). Talhinhas et al. (2008) developed a protocol for efficient Agrobacterium tumefaciens-mediated transformation (ATMT) of C. acutatum. They used hygromycin B as a selective marker, and growth of C. acutatum isolate 397 was completely inhibited at 250 μg/ ml hygromycin B. Consistent with these results, 200 μg/ml hygromycin B media and 800 μg/ml geneticin media were the optimal concentrations for selecting C. acutatum KC05 transformants in the present study.
Fungal transformation is an essential technology in the study of fungal pathogenicity genes at the molecular level. Several fungal transformation technologies have been developed, and PEG-mediated transformation is a simple and inexpensive method (Mathur and Koncz, 1998). Transformation efficiency can vary depending on the technology used and the organism transformed. Armesto et al. (2012) obtained only 21 transformants from C. gloeosporioides (1 × 107 protoplasts/ml) using PEG-mediated transformation, whereas Talhinhas et al. (2008) obtained 45–156 transformants of C. acutatum using the ATMT protocol. Among the 45–156 transformants, Southern blot analyses revealed that more than 70% contained single copy integration of T-DNA (Talhinhas et al., 2008). Maruthachalam et al. (2008) also performed ATMT for two Colletotrichum species, and Southern blot analyses indicated that about 65% of C. acutatum and 62% of C. falcatum transformants contained single copy integration of T-DNA. In the present study, we obtained an average of 103 transformants from 19 different loci of C. scovillei KC05 using PEG-mediated transformation. Southern blot analyses using 2–5 gene knock-out transformants showed that at least one transformant had single copy integration. Moreover, we selected 19 loci from over 10 scaffolds that covered approximately 84.1% of the entire 52,190,760 bp genome to perform a more distributed target gene knock-out (Han et al., 2016). Our results provide a valuable tool for high-throughput genetic analyses of C. scovillei.
Supplemental Materials
Acknowledgments
This study was supported by Basic Science Research Program through the National Research Foundation of Korea grant (NRF-2017R1D1A1B03029622) funded by the Ministry of Education, Science and Technology, and by a grant (918019-04) from the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Notes
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
