Neofusicoccum ribis Associated with Leaf Blight on Rubber (Hevea brasiliensis) in Peninsular Malaysia
Article information
Abstract
Hevea brasiliensis is a natural source of rubber and an important plantation tree species in Malaysia. Leaf blight disease caused by Fusicoccum substantially reduces the growth and performance of H. brasiliensis. The aim of this study was to use a combination of both morphological characteristics and molecular data to clarify the taxonomic position of the fungus associated with leaf blight disease. Fusicoccum species were isolated from infected leaves collected from plantations at 3 widely separated locations – Selangor, Perak, and Johor states – in Peninsular Malaysia in 2010. All the isolates were identified according to their conidial patterns and DNA sequences generated from internal transcribed spacers (ITS1 and ITS2), the 5.8S rRNA, and an unknown locus (BotF15) containing microsatellite repeats. Based on taxonomic and sequence data, Neofusicoccum ribis was identified as the main cause of leaf blight disease in H. brasiliensis in commercial plantations in Malaysia. A pathogenicity trial on detached leaves further confirmed that N. ribis causes leaf blight disease. N. ribis is an important leaf pathogen, and its detection in Malaysia has important implications for future planting of H. brasiliensis.
Members of the Botryosphaeriaceae family are common endophytes, parasites, and saprophytes of a large variety of plants, including monocotyledons, gymnosperms, and angiosperms (Begoude et al., 2010; Slippers and Wingfield, 2007). The rubber tree (Hevea brasiliensis) is a wild tropical plant belonging to angiosperms of the family Euphorbiaceae (Carnier Dornelas and Martinelli Rodriguez, 2005). This important perennial crop and source of natural rubber is currently often affected by various types of diseases which included leaf blight (LB) disease (Nyaka et al., 2012; Radziah and Chee, 1989).
Historically, the causal agent of LB disease was identified as Fusicoccum sp. based on the morphology of the vegetative and reproductive structures observed on lesion tissues (Radziah and Chee, 1989). More recently, Nyaka et al. (2012) focused on the cultural and morphological characteristics of this fungus in an effort to obtain additional information regarding its biology. Despite such efforts, the clear taxonomic position of the fungus responsible for LB disease in rubber remains to be identified. Thus, the correct identification of pathogenic fungi is necessary to implement the appropriate quarantine decisions and suitable control strategies (Denman et al., 1999).
Recently, the advent of molecular analyses using sequence data of the nuclear ribosomal DNA internal transcribed spacer (ITSI and ITS2) regions have supplemented traditional taxonomic methods, enabling a more precise and rapid identification of species in the Botryosphaeriaceae family that are associated with several hosts worldwide (Begoude et al., 2010; Crous et al., 2006; Pavlic, 2009). In this study, we report the use of a combination of morphological characteristics and molecular data to clarify the taxonomic position of a Botryosphaeriaceae fungus associated with LB disease in the Malaysian rubber plant Hevea brasiliensis. Fungal isolates were subjected to DNA sequence analyses of 2 loci, namely the internal transcribed spacers (ITS1 and ITS2), the 5.8S rRNA gene, and an unknown locus (BotF15) containing microsatellite repeats. Pathogenicity trials were then conducted using conidial suspensions as inoculum to clarify the possible role of the isolated fungus associated with LB disease.
Materials and Methods
Fungal isolates
From July to October 2010, rubber leaves exhibiting typical symptoms of blight were collected in 3 regions – Selangor (Sungai Buloh), Perak (Sungai Biong), and Johor (Segamat, Kota Tinggi) – of Peninsular Malaysia. The climatic conditions in these areas are typically tropical, with a mean annual temperature of 26°C, average humidity of 80%, annual average rainfall between 2000–2500 mm (higher for East Malaysia than Peninsular Malaysia) with the northeast monsoon from October to March (wet period), and a southwest monsoon from May to September (dry period).
Samples were collected from rubber trees representing 7 clones in all 3 locations. Ten leaves were cut per tree, and all the samples were placed in paper bags and transported to the laboratory where they were processed after 1 day. Thin sections (5 mm in diameter) of infected tissues were cut with a sterile scalpel, and their surfaces were disinfected via immersion in a 70% ethanol solution for 1 min, rinsing with sterile distilled water, and drying the tissues on sterile blotting papers (Chmlab group, Spain). The disinfected pieces of leaves were placed in petri dishes containing potato dextrose agar (PDA) supplemented with 1 mg·ml−1 streptomycin to minimize bacterial growth. The plates were incubated at 25°C under continuous white fluorescent light for 3 days. Once the fungal colonies were large enough to be examined, the hyphal tips of each colony were transferred to a new petri dish and were incubated as described above to obtain pure cultures. All the isolates obtained were maintained in the culture collection of the Department of Plant Protection, Faculty of Agriculture, Universiti Putra Malaysia (UPM), Malaysia.
Morphological and cultural characteristics
Among the 33 isolates obtained, two strains originating from each geographical region and that have been proved to be virulent genotypes were used in this study. On the eighth day of growth, colour (using a colour scale) (Rayner, 1970), growth, and the presence or absence of conidiomata were recorded for colonies incubated at 26°C on PDA. To study the formation of pycnidia, the same isolates were cultured on sterile pine. The needles were autoclaved twice at 120 °C for 20 min and then placed on petri dishes containing 5 ml of molten water agar, which served as an adhesive. Petri dishes were incubated as described above for 12–15 days. Three petri dishes were used per isolate and medium, and the experiment was performed twice. The length and width of 50 conidia per isolate were measured at 400× magnification under a light microscope (Nikon Model Eclipse E200). The conidia extracted from pycnidia in water agar and pine needles were used for conidial measurement studies, which provided minimum and maximum sizes. Conidial colours, shapes, and septation were also recorded.
DNA extraction
For each isolate, 6 mycelial plugs of a 10-d-old culture grown at 26°C were transferred into Erlenmeyer flasks containing 50 ml Difco potato dextrose broth (24 g/l). The flasks were shaken at 80 rpm at room temperature (28 ± 2°C) for 5 days by using an orbital shaker. The mycelia were filtered through a layer of cheesecloth and washed twice with distilled water. Dried mycelium (250 mg) was mechanically ground to a fine powder in liquid nitrogen by using a pre-chilled mortar and pestle. DNA extraction was performed using the method described by Moller et al. (1992). The concentration of the resulting DNA was determined using an ND-1000 spectrophotometer (NanoDrop 2000; Thermo Scientific, USA). The original DNA was then diluted in sterile distilled water to a concentration of 1 μg/μl and used in further reactions.
Polymerase chain reaction amplification
In order to completely characterize the isolates, the oligonucleotide primers ITS1 (5-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990) were used to amplify and sequence the internal transcribed spacer regions (ITS1 and ITS2) as well as the complete 5.8S rRNA gene. To obtain additional sequence information, another locus was also sequenced. The unknown locus (BotF15) containing microsatellite repeats was amplified with the primers BOT15 (5′-CTGACTTG TGACGCCGGCTC-3′) and BOT16 (5′-CAACCTGCTCA GCAAG CGAC-3′) (Slippers et al., 2004). To amplify each gene region, polymerase chain reaction (PCR) mixtures consisting of the following components were used: 1 μl of each primer (10 mM); 0.5 μl dNTPs (10 mM); 2 μl of MgCl2 (25 mM); 2.5 μl of 10 mM reaction buffer containing MgCl2 (25 mM), (Fermentas, USA); 0.5 U of Taq polymerase (Vivantis, Shah Alam, Malaysia); 5 μl of DNA; and 12.5 μl of sterile distilled water. The following amplification conditions were used: initial denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. PCR products (5 μl) were run on 2% agarose gels in a 1× TAE buffer solution (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) at 80 V for 45 min at room temperature. The gel was stained with ethidium bromide, and the bands were visualized under UV light and photographed using a gel documentation system (GeneSnap, ver 6.03, Syngene Laboratories). The size of the amplified DNA fragment was determined using molecular weight markers (GeneRuler 100 bp DNA Ladder and 1 kb DNA ladder marker [Fermentas, USA]).
DNA sequencing and analyses
The PCR products were purified using a GeneJET PCR Purification Kit (iNTRON biotechnology, Korea). The resulting amplicons were sequenced in both directions by NKH Biosciences Solutions, Kuala Lumpur, Malaysia, by using an automated sequencer. The nucleotide sequences were read and edited with MEGA version 4.0 (Tamura et al., 2007). The sequences of the strains isolated in this study were compared with those from other species in the Botryosphaeriaceae family that are available in GenBank (National Center for Biotechnology Information database). The sequences were aligned using the computer program MEGA version 4.0. Phylogenetic Analysis Using Parsimony (PAUP) version 4.0b10 (Swofford, 2003) was used to perform phylogenetic analyses for the dataset comprising sequences obtained from this study and with those of closely related species in the N. ribis/parvum species complex available from GenBank in order to determine phylogenetic relationships within individual and combined data sets. Gaps were treated as a fifth character, and all the characters were unordered and of equal weight.
Pathogenicity tests
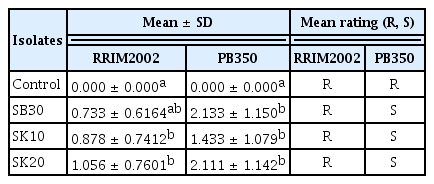
To determine the relative pathogenicity of the fungal isolates, a single rubber leaf was set apart in a 15-cm-diameter petri dish and arranged by using a completely randomized design. One isolate was chosen per location (SB30, SK20, and SK10), and 2 rubber clones (RRIM 2002 and PB350) were used in the trial. For each isolate, a conidial suspension was prepared from 12-d-old cultures grown on PDA plates, and the concentration was adjusted to 3 × 106 spores/ml using a haemocytometer. The leaves were inoculated with micropipettors by placing 10 droplets of the spore suspension onto the abaxial side. The leaves were then placed under continuous fluorescent light (20°C) under humid conditions, with 70–80% relative humidity. Four replicates of leaves were sprayed with a spore suspension of each isolate per rubber clone, and control leaves were inoculated with distilled water. The blighted leaves were assessed 5, 10, and 15 days after inoculation by using a scale (0–4), which represented the following: 0, no visible infection; 1, light lesion around inoculation point lacking spores; 2, large dark lesion without sporulation; 3, large lesion and weak sporulation; and 4, very strong sporulation on the upper and lower surfaces of the leaf. Leaves representing inoculated isolates or uninoculated controls were selected for fungal re-isolations from the edges of the lesion. This experiment was repeated twice for each isolate, and the mean number of lesions per leaflets in each experimental unit (4 leaflets) was computed and expressed as a disease severity index (DSI). The disease index (DI) for each leaf was calculated using the following formula (Ma and Michialides, 2002):
where i is severity (0–4) and Ni is the number of leaves with the severity of i.
The DI for each leaf in each isolate was calculated, and DI < 1 and 1 ≤ DI < 2 were used for classification as resistant (R) and susceptible (S), respectively. Data from all the experiments were analysed using the SPSS statistical package version 17.
Results
Fungal isolation and morphological observations
Fungal isolation from the margins of infected leaves (Fig. 1A) resulted in the recovery of 68 isolates, including 33 resembling members of the Botryosphaeriaceae family, which were fast growers with a white mycelium that turned either dark greenish-grey or greyish within a few days. Overall, 1 type of fungal species belonging to Botryosphaeriaceae was identified by its culture and conidial morphology. These fungi were recovered from all clones commonly grown in Malaysia, including RRIM2024, RRIM901, RRIM2023, RRIM2025, PB350, PB260, and KT39/35. The highest incidence of these isolates was in Johor (16), followed by Perak (10) and Selangor (7). Multiple isolates from Botryosphaeriaceae were obtained from most clones in all of the rubber-growing regions studied.

Symptoms of leaf blight infected by N. ribis. (A) infected rubber leaf; (B) culture plate on PDA; (C) pycnidia produced on PDA plate; (D) conidial morphology (Bar 10 μm).
The characteristics that led to the identification of this isolate were in agreement with previously reported descriptions (Nyaka et al., 2012) shown in Fig. 1B. Colonies were characterized by quick colony growth (16.5–17.0 mm/d), which covered the petri plate after 5 days. In addition, the white fluffy mycelium became slightly yellow or brownish on the undersides after 4 days. By day 5, the centres became light brown and appressed; the white fluffy mycelium was present only at the margins. Furthermore, pycnidia appeared within 7–12 days on PDA (Fig. 1C). Conidia produced on pine needles were initially hyaline and aseptate and became light brown and 1- or 2-septate with age. In addition, the conidia were ellipsoidal with round apices and truncate bases of 16.5–24.0 × 3.5–6.5 μm and a mean L/W ratio of 3.9 μm. No teleomorph structures were formed. The colony and conidial characteristics were typical of Neofusicoccum sp., as described on the Botryosphaeria website (http://www.crem.fct.unl.pt/botryosphaeria_site/).
DNA extraction and PCR amplification
All 33 isolates belonging to the Botryosphaeriaceae family were selected for ITS sequence comparisons to obtain a broad indication of their identities and select isolates for data sets used in the final analyses. We further refined the relationships and identifications of our isolates by performing phylogenetic analyses based on BotF15 sequences. DNA extraction and PCR were successfully performed for both gene regions selected. PCR fragments for the ITS region were determined to be approximately 550 bp in size, whereas those for BOTF15 were approximately 350 bp. BLAST searches against the NCBI (www.blast.ncbi.nlm.nih.gov) database confirmed that the isolates collected represented species in the Botryosphaeraiceae family and indicated that isolates from rubber trees were most closely (99%) related to N. parvum.
DNA sequence analyses
Data sets containing ITS and BotF15 sequences were compiled using sequences obtained from isolates of the Botryosphaeraiceae family in Malaysia and those from the N. ribis/parvum species complex. The ITS data set comprised a total of 18 isolates that is, sequences from 6 isolates (SB30, SB32 SK10, SK20, SB41, SB42) obtained from rubber trees, and 12 sequences from the N. ribis/parvum species complex obtained from the GenBank. The BotF15 data set consisted of 18 isolates, 6 of which were from rubber trees and 12 from the N. ribis/parvum species complex, representing the most complete database of BotF15 sequences in this family (Begoude et al., 2010; Pavlic et al., 2009).
Of the 568 characters in the ITS data set, 475 were constant and 9 were parsimony-informative. Maximum parsimony (MP) analyses generated 100 identical trees (TL = 98, CI = 0.969, RI = 0.786, and RC = 0.762). Isolates from the rubber tree were grouped in a single well-supported clade with N. ribis. During the analyses of the BotF15 data set, the sequences consisted of 364 characters with 356 constants and 7 parsimony-informative characters. MP analyses yielded one of the most parsimonious trees (TL = 8, CI = 1, RI = 1, and RC = 1). The tree generated from using both ITS and BotF15 sequences (data not shown) also clustered the isolates from the rubber tree within the N. ribis clade with strong statistical support.
Concordance among the ITS and BotF15 datasets was confirmed by the calculated incongruence length difference (I = 0), which suggested a lack of conflict between these gene genealogies. The datasets were combined, and a total of 867 bases was generated for the combined ITS and BotF15 datasets. Of these, only 16 characters were parsimony-informative. After heuristic searches, one of the most parsimonious trees of 19 steps (CI = 1, RI = 1, and RC = 1) was obtained (Fig. 2). The consensus tree obtained from the combined analysis of ITS and BotF15 sequences showed that the isolates collected from diseased rubber leaves formed a well-supported clade with N. ribis.

Phylogenetic analysis using Parsimony (PAUP) version 4.0b10 and DNA sequences of N. ribis isolates from three regions in Peninsular Malaysia. The most parsimonious unrooted tree inferred from maximum parsimony analyses of the combined ITS and BotF15 sequence data of the representative taxa of the Botryosphaeriaceae family. The bootstrap support (%) based on 1000 replications is given on the branch. Isolates marked in bold represent those from rubber trees with ITS accession numbers, JX035740, JX035741 (SB32), JX035748 (SK10), JX035764 (SB41), JX035744 (SB42), JX035769 (SK20) and BOT15 accesion numbers JX035770 (SB30), JX035771 (SB32), JX035773 (SK10), JX035774 (SB41), JX035772 (SB42), JX035775 (SB30).
Pathogenicity
All the isolates were capable of infecting leaves, resulting in variable degrees of lesions. The isolates and clones exhibited highly significant differences (p < 0.001) (Table 1). Isolate SK20 produced the largest lesions, followed by isolates SK10 and SB30. Quantitative analysis of these lesions (disease index) indicated a considerable variation in susceptibility to N. ribis. The pathogenicity trials showed the high susceptibility of clone PB350 and the low susceptibility of RRIM2002 (Fig. 3). The leaves showed small light spots 5 day after inoculation. After 10 days, they became slightly brown. By 15 days, undulate margins appeared with pycnidia on the leaf surface and the leaves turned completely brown. Re-isolations from lesions on the inoculated leaves resulted in the recovery of the inoculated fungi.
Discussion
This is the first extensive study on pathogenic fungal strains that induce LB disease in rubber in Malaysia. From all the isolates collected, only 1 species of the Botryosphaeriaceae family was identified on the basis of the morphometric characteristics and analyses of ITS and BotF15 sequences, that is, N. ribis. Based on the virulence demonstrated by isolates of N. ribis in the present study, their presence could possibly explain the recent increase in the frequency of LB disease in commercial rubber plantations in Malaysia.
Conidial colour, size, and morphology coupled with the colony growth and physiognomy on agar plates appear to be the most reliable phenotypic criteria for species identification (Nyaka et al., 2012; Radziah and Chee, 1989). However, such identification in the Botryosphaeriaceae family is complicated due to the confusing taxonomy of anamorphs, and some characteristics thought to be informative at the species level (e.g., conidial pigmentation, septation, and stromata) showed extensive plasticity (Jacobs and Rehner, 1998). Thus, the integration of molecular analyses with traditional phenotypic methods for fungal identification can significantly increase the specificity and decrease the turnaround time for the identification of important fungi (Bagyalakshami et al., 2008). In the present study, the phylogenetic relationships of members of the Botryosphaeriaceae family from infected rubber leaves were determined using individual and combined sequence datasets of the ITS and BotF15 gene regions; consequently, all the isolates were clustered into 1 group with N. ribis. The placement of these isolates within the clade of N. ribis is further supported by their morphological features, which are in accordance with previously reported morphological descriptions (Crous et al., 2006; Pavlic, 2007; Radziah, 1989).
N. ribis and N. parvum are closely related species that formed a species complex within the Botryosphaeriaceae family (Crous et al., 2006). These fungi are known to have both sexual (teleomorph) and asexual (anamorph) stages in their life cycle but are most commonly encountered as anamorphs. The mechanism underlying the sexual reproduction of these species remains to be explored, and little is known regarding their mating strategies (Pavlic et al., 2009). Isolates of N. ribis were initially separated from its sister species N. parvum, based on the similarities between the sequence data of multiple gene regions and PCR-RFLP analyses and were treated as N. ribis sensu lato (Slippers et al., 2004). Recently, 4 species, that is, N. batangarum, N. cordaticola, N. kwambonambiense, and N. umdonicola, were identified in N. ribis sensu lato complex, based on multiple gene genealogies (Begoude et al., 2010; Pavlic et al., 2007).
For many years, the taxonomy of N. ribis and its closely related species has been confusing due to the overlapping morphological characteristics used for their original descriptions. N. ribis was originally described from Ribes spp. in New York, USA, as Botryosphaeria ribis (Grossenbacher and Duggar, 1911). This species was subsequently identified as a pathogen on numerous woody hosts worldwide (Mohali et al., 2007; Pavlic et al., 2007; Rodas et al., 2009; Slippers et al., 2004). In Malaysia, this pathogen was first reported in early 1987 when it was isolated from rubber leaves showing symptoms resembling anthracnose in the state of Johore (Radziah and Chee, 1989). Recently, the disease has spread to 2 other states (Selangor and Perak), which is most likely due to the cultivation of rubber trees in new localities (Nyaka et al., 2012) and environmental factors (The Malaysian Meteorological Department [http://www.met.gov.my]). The temperature in Malaysia is usually 28–35°C, and the transpiration process lowers canopy temperatures (23–24°C). Consequently, it is thought that Hevea trees would be unable to escape N. ribis infection.
The inoculation trials performed in this study showed that all the isolates tested were pathogenic to the young leaves of rubber trees. However, the virulence of these isolates varied with each clone. These results and the consistent association of N. ribis with the disease symptoms seen on rubber leaves suggest that this fungus is a pathogen of H. brasiliensis. Studies on the factors that are related to the susceptibility and resistance of the rubber tree to Fusicoccum leaf disease will provide further insights on the presence of this pathogen on H. brasiliensis leaves.
Acknowledgements
We thank the Malaysian Rubber Board for permission and assistance to conduct this study, as well as Datuk Dr. Abdul Aziz S.A. Kadir, the Secretary General of the International Rubber Research and Development Board (IRRDB), and Dr. Ismail Hashim for their support. We are grateful to Dr Begoude Didier at IRAD Cameroon for his assistance and guidance, and we also thank the Plant Protection Department at the Faculty of Agriculture UPM for logistical support and also thank their staff members Mr. Shamsudin Bojang, Mr. Encik Mohamed Nazri Abdul Rahman, and Mrs. Asmalina Abu Bakar.