The First Identified Citrus tristeza virus Isolate of Turkey Contains a Mixture of Mild and Severe Strains
Article information
Abstract
The presence of Citrus tristeza virus (CTV) has previously been reported in citrus growing regions of Turkey. All serologically and biologically characterized isolates including Iğdır, which was the first identified CTV isolates from Turkey, were considered mild isolates. In this study, molecular characteristics of the Iğdır isolate were determined by different methods. Analysis of the Iğdır isolate by western blot and BD-RT-PCR assays showed the presence of MCA13 epitope, predominantly found in severe isolates, in the Iğdır isolate revealing that it contains a severe component. For further characterization, the coat protein (CP) and the RNA-dependent RNA polymerase (RdRp) genes representing the 3′ and 5′ half of CTV genome, respectively, were amplified from dsRNA by RT-PCR. Both genes were cloned separately and two clones for each gene were sequenced. Comparisons of nucleotide and deduced amino acid sequences showed that while two CP gene sequences were identical, two RdRp clones showed only 90% and 91% sequence identity in their nucleotide and amino acid sequences, respectively, suggesting a mixed infection with different strains. Phylogenetic analyses of the CP and RdRp genes of Iğdır isolate with previously characterized CTV isolates from different citrus growing regions showed that the CP gene was clustered with NZRB-TH30, a resistance breaking isolate from New Zealand, clearly showing the presence of severe component. Furthermore, two different clones of the RdRp gene were clustered separately with different CTV isolates with a diverse biological activity. While the RdRp-1 was clustered with T30 and T385, two well-characterized mild isolates from Florida and Spain, respectively, the RdRp-2 was most closely related to NZRB-G90 and NZRB-TH30, two well-characterized resistance breaking and stem pitting (SP) isolates from New Zealand confirming the mixed infection. These results clearly demonstrated that the Iğdır isolate, which was previously described as biologically a mild isolate, actually contains a mixture of mild and severe strains.
Tristeza disease caused by Citrus tristeza virus (CTV), is one of the most destructive and economically important viral diseases limiting commercial citrus production worldwide (Bar-Joseph et al., 1989; Rocha-Pena et al., 1995). The disease is now distributed in almost all citrus growing regions of the world including Turkey (Moreno et al., 2008; Rocha-Pena et al., 1995). CTV is able to infect most species, varieties and hybrids of Citrus as well as some close relatives of Citrus (Muller and Garnsey, 1984). While most commercial Citrus varieties are sensitive to CTV, a citrus relative, Poncirus trifoliata, and some of its hybrids are resistant to this virus. Therefore, they were used as rootstock in some Citrus-growing regions including the Aegean and the Black Sea regions of Turkey. However, some isolates of CTV breaking the resistance in P. trifoliata were recently identified in New Zealand (Harper et al. 2009; Harper et al., 2010).
CTV is transmitted readily by grafting and several aphid species including Toxoptera citricida, Aphis gossypii and Aphis spiraecola are able to transmit the virus in a semi-persistent manner in nature (Roistacher and Bar-Joseph, 1987). While T. citricida is the most efficient vector of CTV (Yokomi et al., 1994) and widely distributed in many Citrus-growing regions, A. gossypii, acts as a major vector, in some regions including Turkey where T. citricida is still absent. The recent detection and spread of T. citricida in Portugal and Spain (Ilharco et al., 2005) has become a significant concern for many Mediterranean countries such as Turkey where the majority of citrus are grown on CTV-sensitive sour orange rootstocks.
CTV is a complex virus consisting of various strains causing a variety of symptoms in different Citrus hosts. Based on the symptoms inflicted on Citrus scions and rootstocks, CTV strains are divided into five major groups including mild (M), vein clearing in Mexican lime, seedling yellows (SY) in sour orange, lemon or grapefruit, quick decline (QD) in all Citrus species grafted on sour orange rootstock, and stem pitting on grapefruit (SP-G) and on sweet orange (SP-O) regardless of the rootstock. Symptoms of CTV isolates are determined experimentally using a previously established standardized set of Citrus indicator plants (Garnsey et al., 1987).
CTV is a single-stranded positive-sense RNA virus belonging to the genus Closterovirus in the Closteroviridae family. The long thread-like, flexuous, filamentous particles of CTV are about 2000 nm by 11 nm (Bar-Joseph et al., 1979; Bar-Joseph and Lee, 1990) and consist of one single-stranded positive-sense RNA molecule encapsidated with the major capsid protein (CP) and minor CP (Febres et al., 1996). Sequencing of the complete genome of CTV isolates from different geographical origins and with various biological properties confirmed that CTV has one of the largest plant virus genomes ranging from 19226 to 19306 nt. The genome of CTV was organized into 12 open reading frames (ORF) potentially encoding 17 protein products, plus the 3′ and 5′ untranslated regions (UTRs) (Albiach-Marti et al., 2000; Harper et al., 2009; Harper et al., 2010; Karasev et al., 1995; Mawassi et al., 1996; Pappu et al., 1994; Suastika et al., 2001; Vives et al., 1999; Yang et al., 1999). The 25 kDa protein is the major capsid protein (Sekiya et al., 1991) encapsidating about 95% of the CTV genome. Therefore, the CP gene has been used as target for various methods developed for detection of CTV and strain identification including ELISA, western blot, peptide mapping, RT-PCR, hybridization and real-time RT-PCR. In addition the CP genes of a large number of CTV isolates from different geographical regions and biological properties have been cloned and sequenced (Mawassi et al 1993; Pappu, et al., 1993; Roy et al., 2003). Since a correlation between geographical origin and the biological characteristics of CTV isolates and their CP gene sequences was observed (Herrera-Isidrón et al., 2009), sequencing and the phylogenetic analysis of the CP gene was generally used for identification of newly identified and biologically uncharacterized CTV isolates (Niblett et al., 2000; Nolasco et al., 2009).
In Turkey, CTV was first detected symptomatically around Adana by Norman in 1963 and in Aegean Region by Özalp and Azari in 1967 (Baloglu and Birisik, 2009; Yılmaz, 1999). The presence of CTV was demonstrated experimentally by serological and biological assays (Baloglu, 1988) and the first Turkish CTV isolate, Iğdır, was identified (Baloglu, 1988). Since then Iğdır and a limited number of other CTV isolates from Eastern Mediterranean region were characterized by serological and biological assays and these studies indicated that only mild isolates were present in Turkey (Baloğlu 1988; Korkmaz, 2002; Yılmaz and Baloglu, 1998). After the detection of CTV in the Eastern Black Sea region by ELISA and RT-PCR (Korkmaz et al., 2006), the presence of CTV in all citrus growing regions of Turkey was confirmed. Therefore, the presence and distribution of CTV isolates in different citrus growing regions of Turkey is largely known and their serological and biological properties are determined to some extent. However, there is no sequence information available for Turkish CTV isolates and their molecular characteristics are completely unknown. In this study, molecular characteristics of the first identified CTV isolate, Iğdır, was determined by molecular tests used for strain identification and sequencing and phylogenetic analysis of the CP and the RdRp genes.
Materials and Methods
Virus isolate
The Iğdır isolate of CTV originally obtained from a sweet orange tree grafted on sour orange rootstock in a commercial orchard in Iğdır village of Mersin, Province located in the Eastern Mediterranean region of Turkey was used in this study. The isolate was originally found in the 1980s, identified by ELISA (Baloğlu 1988). It was later grafted and maintained on Mexican lime to date. Tissue samples of Mexican lime grafted with CTV Iğdır isolate were obtained from the Western Mediterranean Agricultural Research Institute (BATEM) in Antalya, Turkey where it is currently maintained as a reference isolate in a greenhouse. The samples of T30 and T36 used as mild and severe strains, respectively, were kindly provided by Richard F. Lee of USDA-ARS National Clonal Germplasm Repository for Citrus and Dates.
Oligonucleotide primers
The primers specific to the CP gene were designed based on the conserved sequences at the 5′ end (BC24 5′-ATGGACGACGAAACAAAGAA-3′) and 3′ end (BC25 5′-TCAACGTGTG TTGAATTTCC-3′) of the CTV CP gene. Since the 5′ and 3′ ends of the RdRp gene were not conserved the primers specific to RdRp gene were designed based on conserved regions of located upstream (BC42 5′-CCTACTGAATATAAGGGTAG-3′) and down-stream (BC43 5′-CTCGCGAAGGCAAACAT-3′) of the RdRp gene. The primers for the bidirectional PCR were previously reported (Çevik et al., 1996).
Amplification of the CP and RdRp genes by RT-PCR
dsRNA was isolated using a previously reported dsRNA isolation method (Morris and Dodds, 1979) with minor modification. The CP and RdRp genes of the Iğdır isolate were amplified from dsRNA by a two-step reverse transcription-polymerase chain reaction (RT-PCR) assay using AMV reverse transcriptase and Pfu DNA polymerase (Promega, USA). First, 5 ml of dsRNA was denatured at 95ºC for 5 min and quickly chilled on ice. cDNA was synthesized from denatured dsRNA in 20 μl mixture containing 1X AMV first strand buffer (50 mM Tris HCl pH 8.3, 75 mM KCl, 3 mM MgCl2), 20 units RNAsin (Promega, USA), 0.5 mM dNTPs, 20 units AMV reverse transcriptase (Promega, USA) and 20 pmol random hexamers using the MJ Mini thermal cycler PTC1148 (Bio-Rad, USA) programmed at 30ºC for 10 min. followed by 42ºC for 60 min and 75ºC for 15 min.
PCR was conducted in 50 μl reaction mixture containing 1X Pfu reaction buffer [20 mM Tris-HCl (pH 8.8 at 25°C), 10 mM (NH4)2SO4, 100 mM KCl, 1% (v/v) Triton X-100, 1 mg/ml BSA and 20 mM MgSO4], 0.2 mM dNTPs, 2.5 unit Pfu DNA polymerase (Promega, USA) 5 μl of cDNA and 20 pmol of primer specific to CP (BC24 and 25) and RdRp (BC42 and BC43) genes. PCR was performed in the MJ Mini thermal cycler PTC1148 (Bio-Rad, USA) programmed for initial denaturation at 94°C for 3 min and then, 40 cycles of denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and primer extension at 72°C for 1 min for the CP gene and 2 min for the RdRp gene followed by final extension at 72°C for 5 min. PCR products were separated in 1% agarose gel with 100 bp DNA size marker, stained with ethidium bromide and visualized and analyzed by Doc-It system (UVP, England).
Bidirectional RT-PCR (BD-RT-PCR) assay
Bidirectional-RT-PCR (BD-RT-PCR) method was develop to differentially amplify 300 bp or 400 bp fragment along with full-length CP gene of isolates with or without the MCA13 epitope, respectively, for strain identification (Çevik et al., 1996). BD-RT-PCR was performed as previously described to determine if the CP gene Iğdır isolate contains MCA13 epitope predominantly associated with isolates considered as severe. PCR products were separated in 1.5% agarose gel and analyzed as described in the previous section.
Western blot analysis
Bark tissue from Mexican limes seedlings infected with the Iğdır isolate was ground to powder in liquid nitrogen using a mortar and passel. About 200 mg ground tissue was homogenized in 0.5 ml 2X extraction buffer containing 0.125 M Tris-HCl at pH 6.8, 4% SDS, 20% glycerol, and 10% 2-mercaptoethanol. The homogenate was incubated at 95ºC for 5 min, cooled down to room temperature and briefly centrifuged to separate cellular debris. Supernatant was loaded onto a 12% SDS polyacrylamide gel and total protein was separated by electrophoresis, and transferred to a nitrocellulose membrane (Stratagene, USA) using a Mini Transblot (Bio-Rad, USA). The membrane was probed with the MCA13 monoclonal antibody (Permar et al., 1990) (kindly provided by Richard F. Lee), followed by rabbit anti-mouse IgG conjugated with alkaline phosphatase (Promega, USA). The presence of the 25 kDa major CP was detected colorimetrically with a p-nitrophenol phosphate substrate (Sigma, USA).
Cloning of the CP and RdRp genes
The CP and RdRp genes amplified by RT-PCR from Iğdır isolate were separated in 1% agarose gel, appropriate bands were excised from the gel and purified using the Qiaquick gel purification kit (Qigene, Germany) according to the manufacturer’s instructions. For making compatible ends for T-A cloning an adenine (A) overhang was added to the 3′ ends of the purified DNA by Taq DNA polymerase by A-tailing method. A-tailed PCR products (5 μl) were ligated into the pGEM-Teasy plasmid vector (Promega, USA) at 4°C for 16 hr using T4 DNA ligase and 1X ligation buffer (0.05 M Tris-HCl pH 8.0, 0.01 M MgCl2 1 mM ATP and 50 μg/ml bovine serum albumin). Competent cells of Escherichia coli strain JM109 were transformed with ligation mixture by heat shock at 42°C for 1 min. Transformed cells were plated and grown in LB medium (1% bacto-tryptone, 0.5% bacto-yeast extract, 0.5% NaCl) containing 100 μg/μl ampicillin, 25 μg/ml 5-bromo-4-chloro-Indoly-β-D-galactoside (X-Gal) and 0.1 M isopropyl-beta-thio galactopyranoside (IPTG) at 37°C 16 hr. Recombinant colonies carrying pGEM-Teasy plasmid vector with the CP or RdRp genes were selected by blue/white screening on LB medium according to the manufacturer’s instructions. For each sample, at least five white colonies were screened by colony PCR and at least two colonies carrying the pGEM-T Easy plasmid with the CP and RdRp genes were identified. Plasmids were isolated from these colonies and the presence of the CP and RdRp genes in the plasmids were confirmed by EcoRI digestion.
Sequencing and sequence analysis of the CP and RdRp genes
The cloned CP and RdRp genes were sequenced in both directions by automated cycle sequencing using M13 forward and reverse primers. The sequences of the CP and RdRp genes were assembled and analyzed using Vector NTI Suite program (Invitrogen, USA). The sequences were compared with each other and other CP and RdRp gene sequences in the GenBank. Multiple sequence alignments of the full-length amino acid sequences of the CP and the RdRp genes were conducted by the AlignX Module of Vector NTI Suite. The phylogenetic analysis was performed by Clustal X2 program using neighbor joining algorithm and tested by a bootstrap analysis with 1000 replications. Constructed phylogenetic trees were visualized by TreeView program.
Results
Characterization of the Iğdır isolate
Previously identified Iğdır isolate maintained in Mexican lime as reference mild isolate of CTV was tested by Western blot with MCA13 antibody and the BD-RT-PCR developed based on the MCA13 epitope in CP. Therefore, Western blot was used to determine if the Iğdır isolate react with MCA13 antibody, which is predominantly used for identification of severe, especially QD, strain of CTV. Western blot analysis of the Iğdır isolate was conducted along with T30 and T36, reference for mild and severe strains, respectively. Western blot analysis with MCA13 antibody showed that while T30 did not react with MCA13, T36 and the Iğdır isolate both reacted with MCA13 and a 25 kDa protein corresponding to the CP was detected in both samples. The result showed that the Iğdır isolate previously characterized as mild contains the MCA13 epitope associated with severe isolates (Fig. 1A).

Detection of MCA13 epitope, which is predominantly found in severe strains, in Iğdır isolates of CTV using Western blot (A) and BD-RT-PCR (B) assays. M: DNA or protein size marker; T30: a mild isolate from Florida do not contain the MCA13 epitope; T36: a decline-inducing severe isolate from Florida containing the MCA13 epitope.
The BD-RT-PCR method previously developed for strain identification based on a single nucleotide change in the MCA13 epitope of the CP gene was used to confirm the Western blot results, suggesting that the Iğdır isolate may contain a severe component. The BD-RT-PCR assay was conducted for the Iğdır isolate along with T30 and T36, reference isolates for mild and severe strains, respectively. Using the BD-RT-PCR method, a full-length CPG (700 bp) and a 400 bp fragment specific to mild strains were produced from the mild strain T30 (Fig. 2B). On the other hand about 700 bp fragment corresponding to the full-length CP gene as well as a 300 bp fragment specific to MCA13 positive severe strains were amplified from the severe QD strain T36 and the Iğdır isolate (Fig. 1B). No DNA fragment was amplified from the healthy Mexican lime samples used as negative control. The results demonstrated that the BD-RT-PCR method worked properly and clearly showed that the CP of the Iğdır isolate contains the MCA13 epitope predominantly found in severe strains of CTV. These findings supported the Western blot data and confirmed that the Iğdır isolate contains the MCA13 epitope sequence associated with severe isolates.
Cloning the CP and RdRp genes
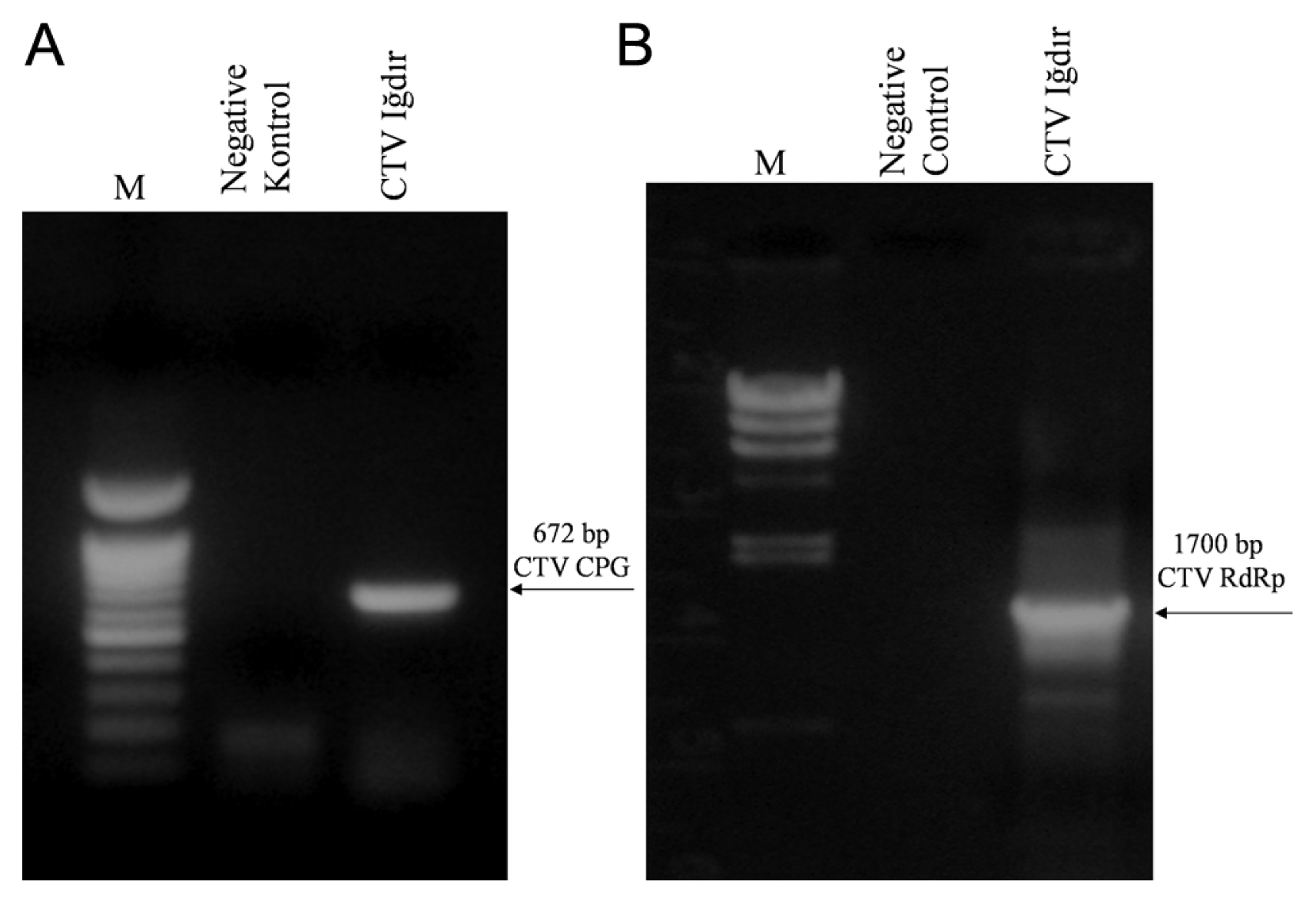
RT-PCR tests were conducted for amplification of the CP and RdRp genes of Iğdır isolate of CTV. While DNA fragments of about 700 bp and 1700 bp corresponding to the CP and the RdRp genes, respectively, was amplified by RT-PCR from cDNA synthesized from the dsRNA isolated from Mexican lime infected with the Iğdır isolate of CTV (Fig. 2), no DNA was amplified from the dsRNA isolated from uninoculated Mexican lime (Fig. 2). The results showed that Mexican lime used as maintenance host still contained the Iğdır isolate of CTV and primers designed based on the CP and RdRp sequences were suitable for amplification of both genes. Therefore, amplification of the CP and RdRp genes enable molecular characterization of the Iğdır isolate using two different genomic regions.
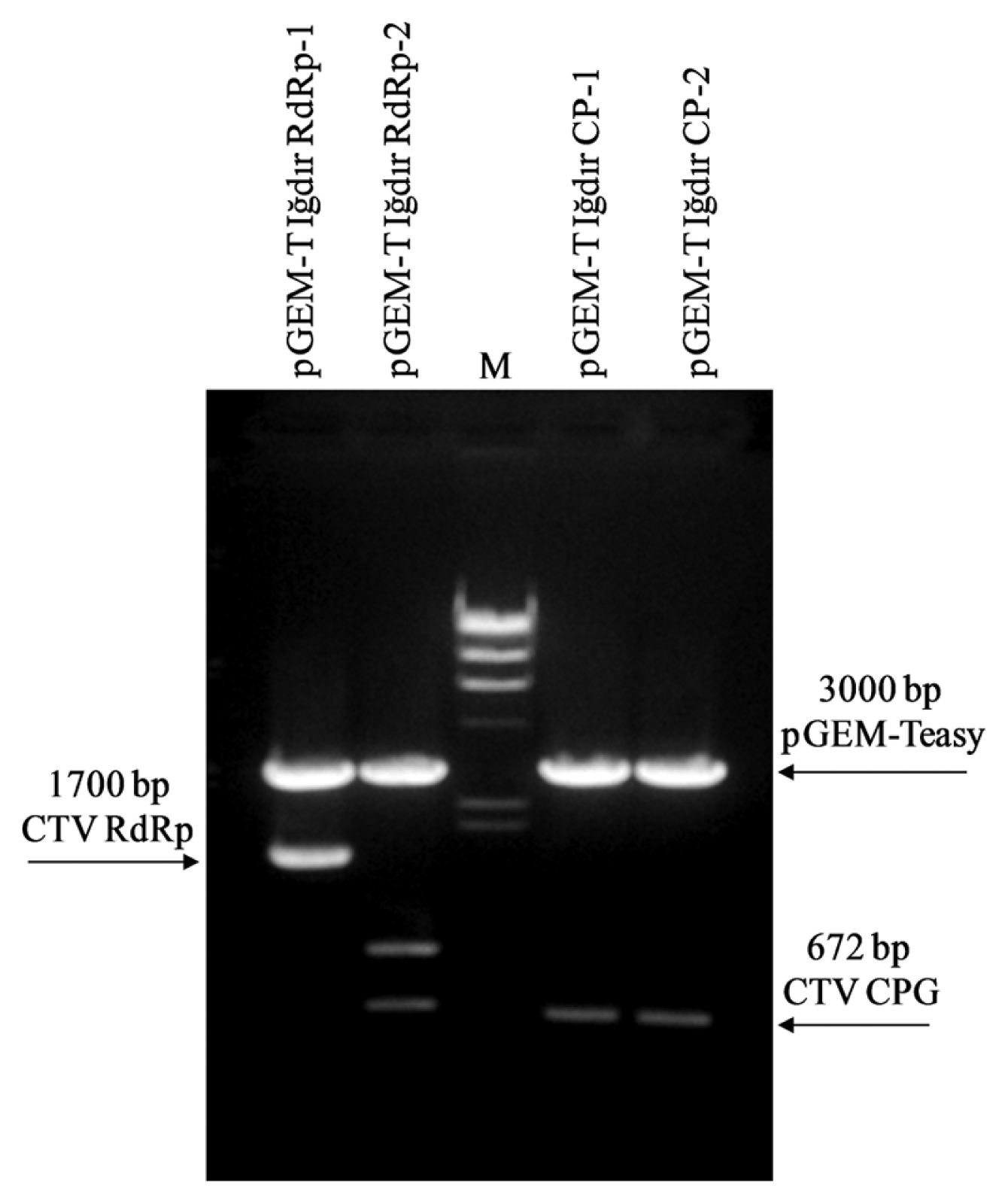
The CP and RdRp genes were cloned in to pGEM-Teasy plasmid vector using T-A cloning method. A number of recombinant colonies potentially carrying the CP and RdRp genes were obtained in initial blue-white screening. At least 10 colonies for each gene were selected and tested by a colony PCR and the presence of the CP and RdRp gene was confirmed in majority of these colonies. Two colonies carrying the pGEM-T easy plasmid with the CP and RdRp genes designated as pGEM-T Iğdır CP-1, pGEM-T Iğdır CP-2, pGEM-T Iğdır RdRp-1 and pGEM-T Iğdır RdRp-2 were selected and plasmid was purified and digested with EcoRI. Digestion of pGEM-Teasy plasmids carrying the CP gene revealed an about 750 bp band containing the CP gene and 3000 bp band of pGEM-Teasy plasmid in both clones (Fig. 3). On the other hand, the digestion of the pGEM-Teasy plasmids carrying the RdRp gene produced different pattern in two different clones. While digestion of pGEM-T Iğdır RdRp-1 clone produced two bands of about 1700 bp and 3000 bp corresponding to RdRp gene and pGEM-Teasy respectively, the other clone produced three different bands. The 3000 bp corresponded pGEM-Teasy vector, the other two fragments (700 and 1000 bp) together corresponded to the RdRp gene suggesting that it has an internal EcoRI site (Fig. 3). The results demonstrated that the CP and RdRp genes were successfully cloned and two different clones for each gene were obtained. Different restriction patterns produced by EcoRI digest of the RdRp clones suggested that these clones contained some sequence variation and the Iğdır isolate may contain a mixture of different CTV strains.
Sequence analysis of the CP and RdRp genes
The nucleotide sequence of two different clones of the CP (Iğdır CP-1 and CP-2) and the RdRp (Iğdır RdRp-1 and RdRp-2) genes were determined and submitted to the GenBank database under the accession KC349866, KC349867, KC349868 and KC349869. The nucleotide and the deduced amino acid sequences of two different clones of the CP gene from the Iğdır isolate were 100% identical (Fig. 4A, Table 1). However, two different clones of the RdRp gene from the Iğdır isolate shared only 90% nucleotide sequence identity and only 91% amino acid sequence identity with each other (Fig. 4A, Table 2). The amino acid sequence comparisons of different clones of the RdRp and CP genes of the Iğdır isolates is shown in Fig. 4. These results showed that while two clones of the CP genes were identical, the RdRp clones were different indicating that two RdRp clones were derived from mixed infection of more than one isolate. To determine the phylogenetic relationships and possible biological activity of the potential isolates in the mixed infection, the deduced amino acid sequences of the CP and RdRp genes from the Iğdır isolate were compared with respective genes from well-characterized CTV isolates from other Citrus growing regions of the world available in the GenBank databases.
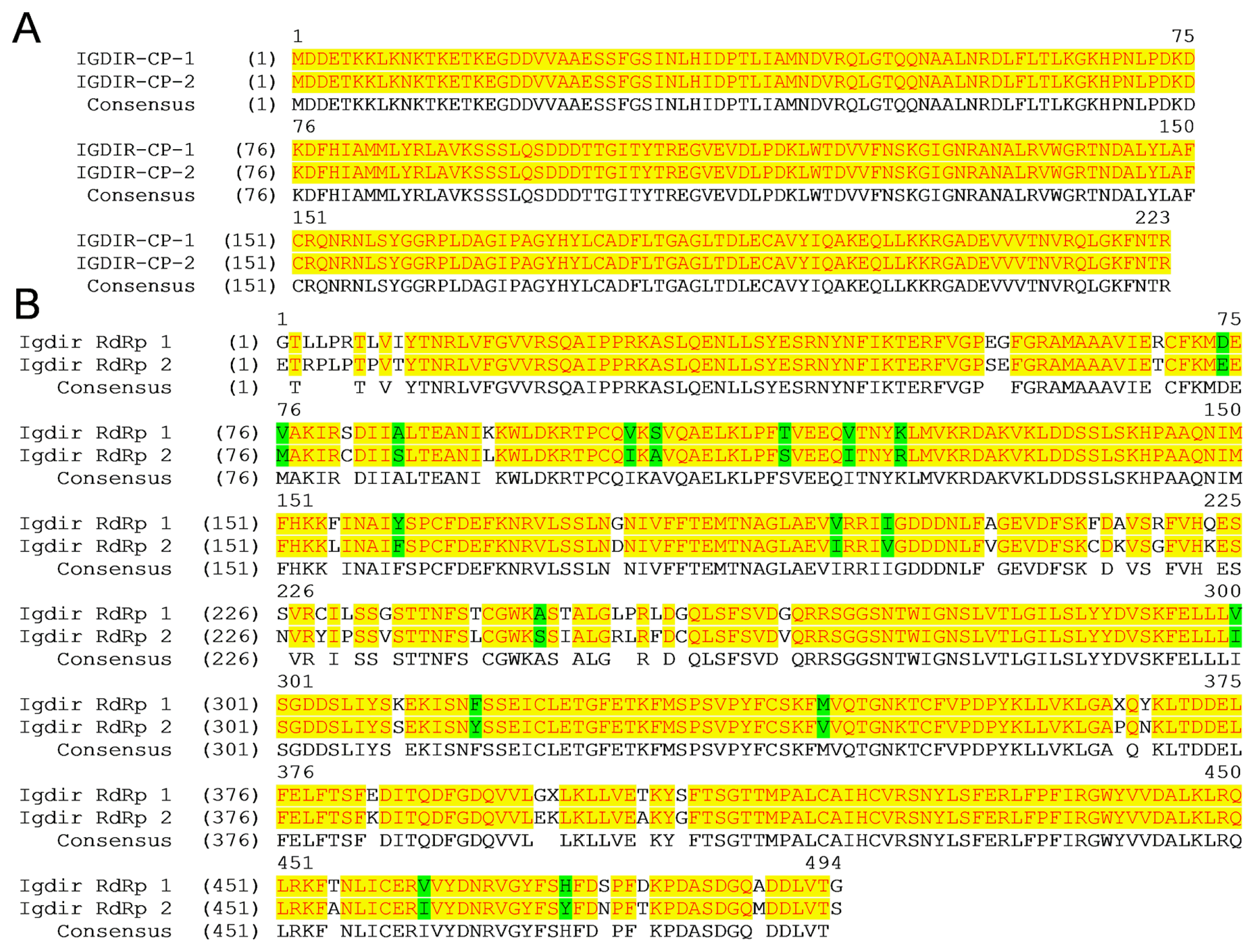
Amino acid sequence alignment of two different clones of the CP (A) and the RdRp (B) genes of Iğdır isolate. The positions of amino acids are indicated above the sequence. The identical and similar amino acids are highlighted in yellow and green, respectively. The GenBank accession numbers for the Iğdır CP-1, CP-2, RdRp-1 and RdRp-2 are KC349866, KC349867, KC349868 and KC349869, respectively.

Percent amino acid sequence identity of the RdRp gene of Iğdır isolates with other CTV isolates from different Citrus-growing regions of the world

Percent sequence identity of the CP gene of Iğdır isolates with other CTV isolates from different Citrus-growing regions of the world
Multiple alignment and comparison of the CP genes revealed that the Iğdır isolate shared 95 to 98% amino acid sequence identity to other CTV isolates. With 98% sequence identity the CP gene of the Iğdır isolate was most similar with the CP gene of NZRB-TH30 (Table 1), a resistance breaking amino acid isolate from New Zealand (Harper et al., 2009; Harper et al., 2010). This isolate is able to break the CTV resistance and replicate in P. trifoliata and also cause severe stem pitting on sweet orange. Interestingly, the CP gene of the Iğdır isolate was the least similar to that of two well- characterized mild isolates, T30 from Florida and T385 from Spain with 95% sequence identity (Table 1). The results clearly demonstrated that the Iğdır isolates contains a severe component and both clones of the CP gene were derived from this severe component of the mixed infection.
On the other hand, multiple alignment and comparison of deduced amino acid sequences of two different clones of the RdRp from Iğdır shared different sequence identity with the RdRp of other CTV isolates. While the RdRp-1 showed 88 to 97 % identity, the RdRp-2 shared 86 to 95% amino acid sequence identity with RdRps from other CTV isolates. The RdRp-1 was 95% identical to five isolates with different biological characteristics, but it was most similar to T30 and T385, two well-characterized mild isolates from Florida and Spain, respectively (Table 2). With 86 and 88% identity it was the most different from a QD isolates from Mexico and QAHA from Egypt, respectively (Table 2). In contrast, with 96 and 97% identity the RdRp-2 was most similar to NZRB-G90 and NZRB-TH30 two characterized RB and SP causing isolates from New Zealand (Table 2). It was the least similar to an orange SP isolate NZ-B18 from New Zealand and a QD isolate from Mexico, with 88 and 89% sequence identity, respectively (Table 2). The result revealed that two different clones of RdRp from the Iğdır isolates show homology with the RdRp of CTV isolates with very different biological activity one being M and the other being the most severe RB isolates of CTV from different citrus growing regions.
Phylogenetic relationships of the Iğdır isolate with previously characterized CTV isolates from different citrus growing regions was determined by comparing the amino acid sequences of the CP and RdRp genes. The phylogenetic analysis of the CP gene revealed four main isolates groups with different biological activity supported by high bootstrap values. The Iğdır isolate were clustered with NZRB-TH30, a RB isolate from New Zealand (Fig. 5). The results showed that two different clones of the CP gene with identical amino acid sequences were closely related with a severe stem pitting isolate enable to break CTV resistance in P. trifoliata confirming that the Iğdır isolate contains a severe component. On the other hand, although the a similar phylogenetic tree was constructed using the RdRp genes of different CTV isolates, two different clones of the RdRp genes of Iğdır isolates were clustered separately with different CTV isolates having diverse biological activity. While the RdRp-1 was clustered with T30 and T385, two well-characterized mild isolates from Florida and Spain, respectively, the RdRp-2 was most similar to NZRB-G90 and NZRB-TH30 two well-characterized RB and SP causing isolates from New Zealand (Fig. 4B). These result clearly demonstrated that the Iğdır isolate biologically described as a mild isolate actually contains mild and severe strains as mixture.

Phylogenetic analyses of the amino acid sequences of the CP (A) and the RdRp (B) genes of Iğdır isolate with other CTV isolates from different citrus growing regions of the world. The origin and biological property of each isolate are shown after the isolate number. M: mild, SY: seedling yellows, QD: quick decline SP: stem pitting. Bootstrap values are indicated on the node of the three. The GenBank accession numbers for the Iğdır CP-1, CP-2, RdRp-1 and RdRp-2 are KC349866, KC349867, KC349868 and KC349869, respectively.
Discussion
Citrus tristeza virus is reportedly present in Turkey for about fifty years but it did not cause any widespread epidemics and economical losses in citrus mainly grown on CTV-sensitive sour orange rootstocks (Baloglu and Birisik, 2009; Yılmaz, 1999). Epidemiology and destructiveness of CTV depends on the type of scion rootstock combination used for production, the presence of vector species and type(s) of strains found in a specific region. Therefore, besides knowing the alternative rootstocks and keeping efficient vectors out, the characterization of strains is very critical for prevention of CTV epidemics in Turkey. Although CTV isolates found in main citrus growing regions of Turkey were biologically and serologically characterized (Baloğlu, 1988; Korkmaz, 2002; Yılmaz and Baloglu, 1998) the molecular characteristics of CTV isolates of Turkey is mainly unknown. Here we report molecular characterization of the first identified CTV isolates, Iğdır, using sequences of the CP and the RdRp coding regions of its genome.
Molecular analysis of the Iğdır isolate with the MCA13 monoclonal antibody (Permar et al., 1990) and bi-directional PCR developed based on the MCA13 epitope (Çevik et al., 1996) revealed that it contains MCA13 epitope indicating the presence of severe strains of CTV. The MCA13 epitope was previously characterized and while MCA13 reactive severe strains contain phenylalanine (F) at position 124 of the CP, mild isolates unrecognized by MCA13 have tyrosine (T) at the same position (Pappu et al., 1993) Sequence analysis of the CP of the Iğdır isolate showed that it contained F at position 124 of the CP confirming the presence of MCA13 epitope. Since presence of the MCA13 epitope predominantly indicates severe strains such as QD, SP and SY, these analyses clearly showed the presence of severe strains in the Iğdır isolate. Although we previously reported the presence of MCA13 reactive strains in Turkey (Korkmaz et al., 2008), finding of this study further confirms and clearly demonstrates the presence of the MCA13 epitope by sequencing of the CP gene, in addition to a positive antibody reaction, in an isolate previously described as mild.
Phylogenetic analysis of amino acid and nucleotide sequences of viral genes has been used for strain identification and molecular characterization. Phylogenetic analyses of CTV isolates were mainly conducted using the CP gene sequences. Comparison of the CP gene of biologically and geographically different CTV isolates revealed that the CP gene sequence correlated with biological activity or geographical origin of isolates (Herrera-Isidrón et al., 2009; Mawassi et al., 1993; Pappu et al., 1993b; Roy et al., 2003). Therefore, it is commonly used for identification of newly discovered strains and genetic relationship among strains of CTV from different citrus growing regions. On the other hand, genome sequences of CTV strains demonstrated that while the 3′ half of the genome is highly conserved, significant sequence variation was observed in the 5′ half of the genome (Albiach-Marti et al., 2000a; Mawassi et al., 1996; Suastika et al., 2001; Vives et al., 1999; Yang et al., 1999). Therefore, two genomic regions, the CP and RdRp, representing respectively the 3′ and 5′ half of the genome were utilized for molecular characterization of Iğdır isolate. Comparison of sequences obtained from two randomly selected clones of the CP and the RdRp genes revealed that two cloned CP genes were identical but the RdRp genes were significantly different from each other suggesting a mixed infection with two different strains. Comparison and phylogenetic analysis of the CP and RdRp genes with respective genes of biologically and geographically different CTV isolates available in the GenBank databases revealed that the CP gene and the one of the RdRp gene were most similar to and grouped with severe SP isolates including NZRB-TH30, a RB isolate from New Zealand (Harper et al., 2010) but the other RdRp gene was more closely related with mild isolates and phylogenetically grouped with mild T30 and T385. This finding suggested that the Iğdır isolate contains both mild and severe components. Similarly, isolates containing mixed infection of mild and severe strains in the same tree was previously reported in different citrus growing regions (Harper et al., 2009; Sentandreu et al., 2006). Particularly one Spanish isolate contain both mild and severe strains which were separated by aphid transmission and host passage and characterized in detail (Ayllon et al., 2006; Sentandreu et al., 2006). Similarly, some CTV isolates from New Zealand were sequenced and genomes of different strains were obtained from the same isolates (Harper et al., 2009). These and other findings clearly showed that mixed infections are common and the component of mixed infection can be separated by aphid vectors experimentally as well as naturally in the field.
Therefore, from epidemiological and control point of view it is important to know the biological, serological and molecular characteristics of CTV isolates in the region. Biological characterization of the Iğdır isolate was previously completed and based on the reaction in indicator plants it was considered a mild isolate (Baloğlu, 1988; Yılmaz and Baloglu, 1998; Korkmaz, 2002). The finding in this study showed that the Iğdır isolate actually contains mixed infections of mild and severe SP isolates. Since the biological indexinğf the Iğdır isolate was done by graft inoculation all components of the mixed infection graft inoculated and strains in the isolate were not separated. Therefore, the mild nature of the Iğdır isolate in the indicator plant can be explained by interaction of different strains and/or cross protection of severe component by the mild strains found together. Determination of this has to await separation of mixed strains by aphid transmission.
Citrus tristeza virus is present in all in main Citrus-growing regions of the Mediterranean coast for many years (Baloğlu, 1988; Yılmaz and Baloglu, 1998; Korkmaz, 2002) and recently reported in other Citrus-growing region of Turkey (Korkmaz et al., 2008). Studies indicated that virus is not spreading constantly from tree to tree but rather found sporadically (Baloğlu, 1988; Yılmaz and Baloglu, 1998; Korkmaz, 2002; Korkmaz et al., 2008). This is probably due to the absence of the efficient vector T. citricida. Current predominant vector is A. gossypii, which unable to transmit CTV efficiently (Roistacher and Bar-Joseph, 1984; 1987). However, possible introduction of T. citricida may change the situation. It has been well documented in Florida that the predominant strain profile was dramatically changed after introduction of T. citricida and severe SP isolates become more prevalent (Halbert et al., 2004).
This study provides serological and molecular data as well as sequence information for the presence of severe strain component in the previously identified Turkish CTV isolate Iğdır, which is known as a mild isolate. Therefore, it is certain that the severe strains of CTV are actually present in citrus producing areas of Turkey. These strains are presumably transmitted but are not separated from mild strains by transmission by A. gossypii in nature in the absence of the most efficient vector, T. citricida. A possible introduction of T. citricida, which is currently found in Spain and Portugal, into Turkey may cause efficient transmission of severe components in so-called mild isolates and may change the predominant strain profile. Newly transmitted predominant severe strains could result in epidemics in citrus growing regions of Turkey where the majority of citrus is grafted on CTV-sensitive sour orange rootstocks. Therefore, the CTV isolates and/or strains found in Turkey should be detected, identified characterized in genetic diversity and epidemiological point of view and CTV resistant rootstocks and scions should be developed in preparation for the inevitable results of T. citricida introduction into Turkey.
Acknowledgments
This study was conducted by a grant from the Scientific and Technological Research Council of Turkey (TÜBİTAK) project number 104O584.