Phylogenetics and Gene Structure Dynamics of Polygalacturonase Genes in Aspergillus and Neurospora crassa
Article information
Abstract
Polygalacturonase (PG) gene is a typical gene family present in eukaryotes. Forty-nine PGs were mined from the genomes of Neurospora crassa and five Aspergillus species. The PGs were classified into 3 clades such as clade 1 for rhamno-PGs, clade 2 for exo-PGs and clade 3 for exo- and endo-PGs, which were further grouped into 13 sub-clades based on the polypeptide sequence similarity. In gene structure analysis, a total of 124 introns were present in 44 genes and five genes lacked introns to give an average of 2.5 introns per gene. Intron phase distribution was 64.5% for phase 0, 21.8% for phase 1, and 13.7% for phase 2, respectively. The introns varied in their sequences and their lengths ranged from 20 bp to 424 bp with an average of 65.9 bp, which is approximately half the size of introns in other fungal genes. There were 29 homologous intron blocks and 26 of those were sub-clade specific. Intron losses were counted in 18 introns in which no obvious phase preference for intron loss was observed. Eighteen introns were placed at novel positions, which is considerably higher than those of plant PGs. In an evolutionary sense both intron loss and gain must have taken place for shaping the current PGs in these fungi. Together with the small intron size, low conservation of homologous intron blocks and higher number of novel introns, PGs of fungal species seem to have recently undergone highly dynamic evolution.
Gene families are present in all eukaryotic genomes and constitute significant portions of their genomes; as much as 15% in humans (Li et al., 2001), 13% in yeast (Wolfe and Shields, 1997), and even higher in angiosperm plants (Horan et al., 2005). Completed genome sequencing projects in model species revealed that duplication at the genome level occurred in early stage of eukaryotic evolution, which was followed by the regional or tandem duplication to account for the increment of copy numbers for gene families (Chapman et al., 2004; Prince and Pickett, 2002). If gene duplication occurred prior to the divergence of eukaryotic phylogenetic lineage, the gene families could spread to the specific lineage by lineage-specific expansion (Lespinet et al., 2002). Even in low eukaryotic species such as yeasts, whole genome duplication and subsequent massive loss of duplicated genes by small deletion were noted by whole genome sequence comparison between Saccharomyces cereviseae and Kluyveromyces awalitii (Kellis et al., 2004).
Interruption of coding sequences by introns is a ubiquitous feature of eukaryotic genes. Despite intensive study for more than 30 years, the origin and evolutionary role(s) of introns are still elusive and not well defined. However, it is evident that they are involved in various cellular and developmental processes via alternate splicing or regulation of gene expression (Lees-Miller et al., 1990; Lynch and Richardson, 2002). Depending on the location of introns within a gene, intron can be defined into three phases: phase-0 between codons, phase-1 between the first and the second nucleotides of a codon, and phase-2 between the second and the third nucleotides of a codon. The phases of an intron are a conservative character of eukaryotic gene structures during evolution, because a variation in intron phase is possible only through simultaneous mutations that alter the 5′ and 3′ ends of the intron in a complementary manner. The distribution of intron phases is also unequal. Typically, phase-0 introns are the most frequent and phase-2 introns are the least (Fedorov et al., 1992; Long et al., 1995; 1998).
There are two contradictory schools of thought on the origin of introns, “intron-early” (Doolittle, 1978; Gilbert, 1978) versus “intron-late” (Palmer and Logsdon, 1991; Sverdlov et al., 2007). While the former theorizes that the current genes were derived from extant genes by exon shuffling prior to the divergence of prokaryotes and eukaryotes, the latter argues that the extant genes were intronless but the introns invaded into the continuous genes recently. However, neither theory is sufficient to explain exclusively the eukaryotic gene evolution. For example, a combination of both processes might have occurred frequently to shape current eukaryotic gene structure (Bon et al., 2003; Mourier and Jeffares, 2003). A synthetic theory of merging both ‘intron early’ and ‘intron late’ theories was proposed to explain the new as well as ancient introns in the current genes (de Souza, 2003; Roy, 2003). Due to the functional redundancy, extra copies of gene families of some paralogs might have undergone purifying selection while others were degenerated by accumulation of evolutionarily neutral or loss-of-function mutations (Prince and Pickett, 2002). Therefore, comparison of gene structures of orthologs and paralogs in gene families among related species might give insights into the intron evolution in eukaryotic genes.
Aspergillus is a filamentous fungal genus with over 185 species (Jones, 2007; Timberlake and Marshall, 1989). It contains important species for genetic model, human health and industry. A. nidulans and A. niger have been studied for eukaryotic cellular physiology and molecular biology by a well-characterized sexual cycle and genetic systems (Coppin et al., 1997; Pontecorvo et al., 1953). A. fumigatus is a serious life-threatening human pathogen (Denning, 1998), and A. oryzae is a beneficial food-industry fungus in the production of sake and soy sauce (Abe et al., 2006). With the importance of human health and industrial impact, whole genome sequencing was done in three species, A. nidulans, A. fumigatus, and A. oryzae (Galagan et al., 2005a; Machida et al., 2005; Nierman et al., 2005) and partially or nearly completed in five species, A. clavatus, A. flavus, A. niger, A. terrus, and A. parasiticus (Jones, 2007). These analyses provide good venues for the evolutionary insights of the fungi by comparative genome analysis (Galagan et al., 2005a; 2005b; Payne et al., 2006; Wortman et al., 2006). Neurospora crassa is also a filamentous fungus and has been used for genetic model species (Reviews in Hynes, 2003, references therein). The completed whole genome sequences of N. crassa are now providing valuable resources in comparative genomics in fungal genetics (Galagan et al., 2003; 2005b).
Polygalacturonase (EC3.2.1.15) is a pectin-digesting enzyme containing a glycoside hydrolase family 28 domain. Polygalacturonase (PGs) genes are present in families in most eukaryotes (Markovic and Janecek, 2001). Polygalacturonase is involved in various developmental processes in plants and pathogenicity in phyopathogenic fungi (Hadfield and Bennett, 1998; Markovic and Janecek, 2001). The current study mines the whole or nearly whole sets of PGs in five aspergilli and N. crassa for analysis of their phylogenetic relationships and intron dynamics in the fungal PGs.
Materials and Methods
PG gene sequence mining.
Nucleotide and polypeptide sequences of the PGs were isolated by BLAST program provided from Aspergillus genome database (http://www.aspgd.org/) using the glycohydrolase 28 domain (pfam00295) sequence of yeast for a query sequence. PGs with truncated sequences were not included in the analysis. The PGs of N. crassa were isolated from BLAST X search at the NCBI (http://www.ncbi.nlm.nih.gov/).
Phylogenetic analysis.
T-coffee program (http://www.ebi.ac.uk/t-coffee/help.html/) was used for polypeptide sequence alignment of PGs. Phylogenetic analysis was carried out using Phylip version 3.69 program (http://evolution.genetics.washington.edu/phylip.html) with a bacterial PG for an out-group by 100 repetitions for confident bootstrapping. A phylogenetic tree was developed by the TreeView PC program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Gene structure analysis.
Gene structures with intron and exon sequences were obtained from the species websites. For intron phase identification, the cDNA sequences were compared with genomic DNA sequences by CLUSTAL W program (http://www.ebi.ac.uk/clustalw/) and confirmed by translation using a program in the Sequence Manipulation Suite website (http://bioinformatics.org/sms/). Intron positions were identified manually after the multiple sequence alignment analysis with the CLUSTAL W program.
Results and Discussions
Mining of the PG genes.
The number of PGs mined was 8 in A. nidulans, 6 in A. terreus, 7 in A. niger, 9 in A. fumigatus, 15 in A. oryzae, 2 in A. flavus, and 2 in N. crassa, respectively. The actual number of PGs in each species may be higher than these numbers because amino acid sequences with higher E-value than > 10−4 against query PG or with truncated glycohydrolase 28 domain less than 70% in coverage were not selected. Also, PGs without either mRNA or genomic DNA sequences were excluded from gene structure analyses. The higher number of PGs may be related to the largest genome size of A. oryzae, among the five aspergilli, as shown in other gene families, such as cytochrome p450 enzymes and nonribosomal peptide synthases, which were 151 and 24 copies in A. oryzae and 14 and 65 in A. fumigatus, respectively (Machida, Asai et al., 2005; Payne, Nierman et al., 2006). By whole genome comparison between A. oryzae, A. fumigatus, and A. nidulans, sequence acquisition was found to increase the genome size in A. oryzae (Galagan et al., 2005a).
Polygalacturonase containing the glycohydrolase family 28 domain is one of the largest glycohydrolases encoded by a gene family in eukaryotes (Markovic and Janecek, 2001). The number of PG copies in Aspergillus is considerably lower than those in Arabidopsis (67 copies) and rice (48 copies) (Yokoyama and Nishitani, 2004; Kim et al., 2006).
Gene families were derived via gene duplication and subsequent regional or segmental duplication, which would ultimately be scattered throughout the genome by genome rearrangement (Lynch and Conery, 2000). If the duplication occurred prior to divergence eukaryotic phylogenetic lineage, as shown in the Cytochrome P450 gene family in four filamentous Ascomycetes fungal species, Fusarium graminearum, Magnaporthe grisea, A. nidulans, and N. crassa (Deng et al., 2007), the genes could spread into lineage-specific expansion. In our analysis of Aspergillus and N. crassa, the PGs showed some clade or sub-clade specific gene structures that support the notion of lineage-specific expansion after the genus diverged. In contrast to with the diploid-prominent organisms such as Arabidopsis and rice, the haploid stage is prominent during the life cycle of fungi. Therefore, the duplicated genes in fungi might have subjected different selection pressure from those of the diploid-prominent species. After undergoing duplication, one of the pair may either degenerate to a pseudogene or acquire a novel function (neo-functionalization) (Prince and Pickett, 2002). Expression of the fungal PGs in the current analysis is obvious since mRNA derived cDNA sequences are available in most of them. Functional redundancy of PGs was noted in plants such as corn (Allen and Lonsdale, 1992) and Arabidopsis (Hadfield and Bennett, 1998) because cell wall modification by polygalacturonases is critical in development of plants. However, pectin is not a major component making up cell wall of fungi. Hyper expression of PGs may be related with virulence of the phytopathogenic fungi by softening the plant cell wall to permit penetration by fungal hyphae during the process of infection (Markovic and Janecek, 2001). Except for A. flavus, the other four aspergilli in the current analysis are not known to be phytopathogenic.
Phylogenetic analysis.
The 49 PGs that were analyzed in this study were separated into three clades, which were also divided into 11 sub-clades, based on the amino acid similarities (Fig. 1). While PGs in clade I were rhamno-PGs, those in clade II were exo-PGs. In clade III, PGs were endo-PGs except for the three PGs of sub-clade III-I. PGs in the PGs in sub-clade III-I, which were placed as the most out-group in clade III, were exo-PGs. Hadfield and Bennett (1998) classified plant PGs into three clades of A, B, and C where clade C was composed exclusively of exo-PGs. In the analysis of diverse origin of PGs from plant, fungus, insect, and bacteria (Markovic and Janecek, 2001), fungal PGs formed two separate clusters, one exclusively with exo-PGs and the other with endo-PGs and rhamno-PGs, in which the latter were closely clustered with insect PGs. The difference of our classification from others a further analysis.

Phylogenetic dendrogram and gene structures of PGs of Aspergillus, N. crassa, and E. coli. In the phylogentic tree, the numbers at the nodes are bootstrap values. Lower than 50 bootstrap values were not shown. In gene structure, exons are filled bars and introns are lines. The lengths of the filled bars are relative sizes of the exons, but the lines for introns are not depicting the intron sizes for illustration. The numbers over the gene structure are the number in nucleotides of exons and introns. The colors showing the numbers in intron size are blue for phase 0, red for phase 1, and green for phase 2, respectively. The clusters in right column are the sub-clades; clades are indicated by Roman numerals.
None of the sub-clades contained solely paralogous genes from a single species, which implies that PGs were diverged before species divergence within Aspergillus took place. Also, the separation of two PGs from N. crassa into separate clades implies that the PG divergence into current clades represents predated divergence of Aspergillus and Neurospora which occurred approximately 300 M (Galagan et al., 2005b; Padovan et al., 2005). Clade I contained seven PGs of A. oryzae, A. terreus, and A. fumigatus, which grouped into two sub-clades where bootstrap value in the node of the two sub-clades was 100, but the bootstrap values in the deepest branch in the sub-clade I-I were below 50. Nineteen PGs of A. oryzae, A. terreus, A. fumigatus and A. nidulans were in the clade II which was subdivided into four sub-clades. The bootstrap values in the nodes of sub-clades were high. However, they were lower than 50 in the nodes tying with the sub-clades. One PG from N. crassa (Ncra957508) was tied with four PGs of Aspergillus with 100 bootstrap value in the sub-clade II-II. In a previous study, five PGs of A. oryzae in this clade (Aory66570 in the sub-clade II-II, Aory54924, A ory63240, Aory56683, Aory61240 in sub-clade II–IV) were grouped with plant PGs (Park et al., 2008). However, they ruled out the possibility for fungal PGs being ancestral to the plant PGs since there were no PGs with intermediate gene structure between plant and fungus found in their study. The clade III was the largest one with 23 PGs from A. oryzae, A. terreus, A. fumigatus, A. nidulan, A. niger, A. flavus and N. crassa. There were five sub-clades of the Aspergillus PGs and the PG from N. crassa was not included in the Aspergillus sub-clades of clade 3.
Aspergillus species have evolved and diverged over 200 million years (Galagan et al., 2005a; 2005b). Whole genome duplication and subsequent gene loss predated the Aspergillus speciation in eukaryotic evolution (Achaz et al., 2001; Kellis et al., 2004). The distribution of PGs from each Aspergillus species in the sub-clades supports concept of duplication of PG predating a Aspergillus speciation. The absence of PGs from A. niger in clades I and II might be due to the underestimation of the PG sequences in the partially finished genome project. Among the seven pairs of the PGs in the deepest branch with 100% bootstrap values, one was a pair of paralogous sets of A. niger (Anig12554 and Anig42809 in sub-clade III-III) and two were orthologous PGs from A. oryzae and A. flavus (Aory58591 and Afla05020 in sub-clade III–IV, Afla05015 and Aory58737 in sub-clade III–V) which taken together support the close species relationship of A. oryzae and A. flavus as also shown by a comparison of large genome sequences (Payne et al., 2006).
Gene structure analysis.
Five of the total 49 PGs analyzed did not carry introns. The number of introns per gene varied from one to seven among the rest of the PGs. The average number of introns per gene was 2.5, which was similar to the number of exons in whole genomes of A. nidulans (3.6), A. fumigatus (2.8), and A. oryzae (2.9) (Galagan et al., 2005a). The average number of introns was 3.7 in clade I, 2.9 in clade II, and 1.8 in clade III, respectively (Table 1). The lengths of introns ranged from 20 to 424 bp with an average of 65.9 bp. Most of the introns were between 31 to 90 bp at a peak of 51 to 60 bp (Fig. 2). Parsch (2003) also noted a narrow range of intron length distribution in 15 orthologous genes of Drosophila. Based on these observations, natural selection seems to have played a role in maintaining intron size in contrast to the selective neutrality of the nucleotide sequence variation in introns in various organisms. The average length of the introns of all genes of A. fumigatus was 112 bp which is similar to the introns of another filamentous fungus Phanerochaete chrysosporium (117 bp) (Martinez et al., 2004). The average length of the introns of Arabidopsis PGs was 100 bp (Torki et al., 2000) and minimal intron length in genes of Drosophila was 61 ± 10 bp (Yu et al., 2002). The reason for the short introns in PGs of Aspergillus is not clear at this moment although a negative correlation was reported between intron length and gene expression level in Caenohabditis elegans and Homo sapiens (Castillo-Davis et al., 2002). Another interesting observation was that two or more introns of the same length within a gene were clustered in 8 PGs (Fig. 1). For example, Aory57693 (I–II) had three introns of 66 bp with different intron phases. Afum742685 (II–III) had four introns of 51 bp with one of phase 1 and other 3 in phase 2. The sequences of these introns were highly variable except of those sequences for proper splicing (Fig. 3). An interesting speculation is a concerted evolution for intron size in the spliceosomal introns, which requires further analyses with more data sets.

Intron phase distribution and average number of introns in each clade of PG genes in Aspergillus and Neurospora species analyzed

Distribution of intron size in PGs of Aspergillus. The numbers in X and Y axis are the number of nucleotides and number of introns, respectively.

Multiple nucleotide sequence alignment of the same length of Aory57693 and Afum742685, respectively. The 5′ (GT) and 3′ (AG) ends of the introns are highlighted with red and the conserved sequences for lauriate structure formation are highlighted with blue where the adenine was red.
Among the 124 introns, 18 introns were present in novel positions in only one PG and the rest were arranged in 29 blocks of homologous intron sets. In sub-clade III-II, the introns of the first (50 bp) of Anid6656, the second (50 bp) of Aory55286, and the second (58 bp) of Anig72931, which were differentiated only by 1 amino acid, were counted as a homologous intron set since the slightly different positions might have been derived from intron “sliding” by clustering algorithms (Stoltzfus et al., 1997). Although the homologous introns corresponded in their positions, their nucleotide sequences and length were somewhat variable. None of the intron blocks was common in all clades. Twenty-six of the 29 homologous intron blocks were sub-clade specific. There were higher numbers of introns retaining their positions in clade III compared to those in clade I and II. Intron position conservation in Aspergillus seemed to be less stringent compared to the plant PGs reported by Park et al. (2008), where numerous introns corresponded in their positions between clades. In the analysis of 446 introns from 108 PGs of plants, they showed 19 homologous intron blocks among which only two homologous intron blocks were sub-clade specific and novel introns were as rare as 3 out of the 446 introns. Therefore, 18 novel introns of the 123 introns in the current study are significantly higher than those of plant PGs. The novel introns should have been derived from recent insertion of intronic sequences. Roy (2004) proposed transposon insertion for the origin of recent intron novelty. However, none of the introns in the PGs of Aspergillus had significant homology with known transposon sequences in BLAST analysis. Intron phase distributions among the 18 novel introns were 8 for phase 0, 7 for phase 1 and 3 for phase 2, respectively, to show no phase preference for the insertion site.
Phase distribution of introns of the 124 PGs was 80 for phase 0 (64.5%), 27 for phase 1 (21.8%), and 17 for phase 2 (13.7%), respectively, which was similar to those of plant PGs (65.5% for phase 0, 19.7% for phase 1, and 14.8% for phase 2) (Park et al., 2008), but dissimilar to those of other eukaryotic genes (50% for phase 0, 30% for phase 1, and 20% for 2) (Fedorov et al., 2002). PGs in clade 1 showed a significantly higher number of phase 1 introns compared to those of clade 2 and 3. Since loss of phase 0 introns does not disrupt the protein coding frame, Fedorov et al. (2002) argued that intron loss might have occurred only in phase 0. In our analysis, intron loss was assumed to have occurred if one PG did not have an intron while more than two other PGs had introns at the corresponding position in a sub-clade (Fig. 4). There were 18 intron losses recorded in this study which were disproportionately distributed over the intron phases; 15 for phase 0, 2 for phase 1, and 1 for phase 2, respectively. Figure 4 is showing the intron gain and loss of sub-clade II–III where only one intron gain (phase 0) is evident SPN/WHN in Anid9045 and the other 7 introns (4 of phase 0, 2 of phase 1, 1 of phase 2) were lost in one or two orthologous genes. Both intron loss and intron gain might have occurred in leading up to the present PGs in Aspergillus and N. crassa, which is congruent with the synthetic theory of merging both ‘intron early’ and ‘intron late’ theories (de Souza, 2003; Roy, 2003).
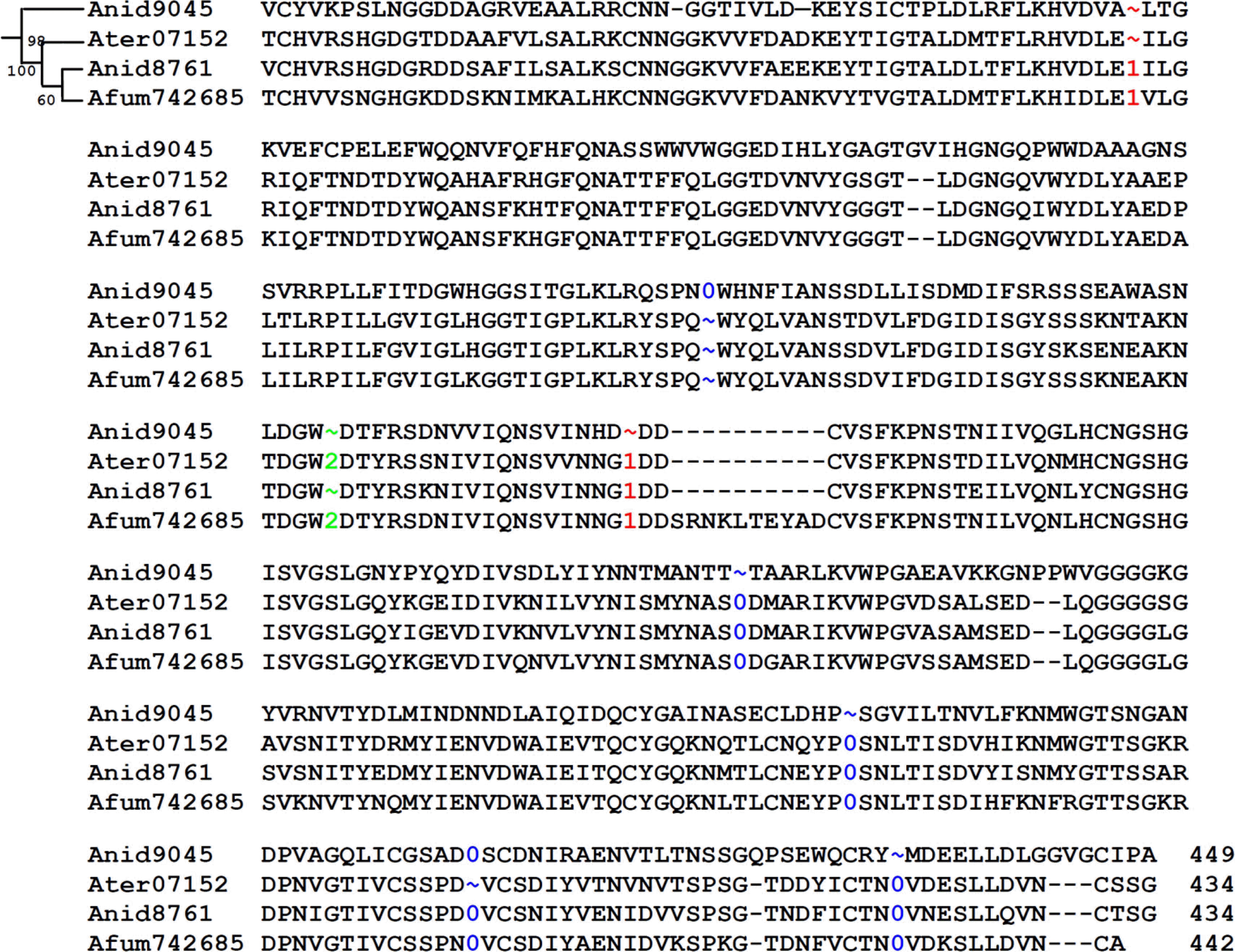
Multiple polypeptide sequence alignment of PGs of clade II–III. The positions of introns were shown by numeric for phase 0, 1, and 2, respectively. Detailed description on the intron loss/gain of the figure is in text.
Overall evolutionary dynamics of gene structures in PGs of Aspergillus and N. crassa with their evidences of small introns, less conservation of intron positions, and higher number of novel introns seemed to be different from plant PGs. The reason for shorter introns of PGs than other fungal genes is not clear at this moment. Although the physiological and developmental roles of PGs are obscure in fungi, their expression is abundant because the cDNA sequences are available in most of the PGs. Elucidation of the cellular and physiological functions of the PGs in fungi will help to understand the role of PGs in phytopathogenicity.
Acknowledgements
Authors appreciate Drs Byron Johnson and George Fedak for critical reading and comments. This work was supported by a research grant from Seoul Women’s University (2012) and an Agenda (PJ007446052011 and PJ907062) from Rural Development Administration, Republic of Korea.