Evaluation of Bt-cotton Genotypes for Resistance to Cotton Leaf Curl Disease under High Inoculum Pressure in the Field and Using Graft Inoculation in Glasshouse
Article information
Abstract
Bt-cotton germplasm, consisting of 75 genotypes was evaluated against cotton leaf curl disease (CLCuD) under high inoculum pressure in the field and using graft inoculation in glasshouse by visual symptom scoring assessments. None of the tested genotype was found disease free under both evaluation tests. Under field conditions in 2011, 3 genotypes were found resistant, 4 moderately resistant, 3 tolerant, 2 moderately susceptible and one susceptible; in 2012, 3 genotypes were tolerant, 7 moderately susceptible, 5 susceptible and 38 highly susceptible; in 2013, one was moderately susceptible and 51 were highly susceptible with varying degree of percent disease index (PDI) and severity index (SI). However, through graft evaluation in glasshouse, none of the graft inoculated plant was symptomless. All tested genotypes showed disease symptoms with SI values ranging between 5.0 and 6.0, and latent period between 12 and 14 days. Of the 75 genotypes evaluated using graft inoculation, 11 were found susceptible with SI values of 5.0 to 5.4 while remaining 64 were highly susceptible with SI values of 5.5 to 6.0. Inoculated plants of all tested genotypes exhibited severe disease symptoms within 10 days after the appearance of initial symptoms. No reduction in SI value was observed until the end of the experiment i.e., 90 days after grafting. Information generated under the present study clearly demonstrates that no sources of resistance to CLCuD are available among the tested Bt-cotton genotypes. So, a breeding programme is needed to introgress the CLCuD-resistance from other resistant sources to agronomically suitable Bt-cotton genotypes.
Cotton (Gossypium hirsutum L.) is one of the most important fibre and non-food cash crops in Pakistan (Khan et al., 2010). It is important for the country’s economy as it ranks at the top as foreign exchange earnings (Manzoor et al., 2013; Wajid et al., 2014). However, for the last more than two decades cotton leaf curl disease (CLCuD) is considered as the major factor for its low productivity in this region. CLCuD is caused by whitefly (Bemisia tabaci Gennadius) transmitted viruses of the genus Begomoviruses (family Geminiviridae) and a satellite DNA β (Akhtar et al., 2014; Manzoor et al., 2013). Since its first epidemic during 1990s, in Pakistan and north western India, at least nine distinct begomovirus species have been found responsible to cause CLCuD (Yousaf et al., 2013). However, during 2001 only a single recombinant begomovirus strain namely Cotton leaf curl Burewala virus (CLCuBuV) was identified in cotton across the Punjab province of Pakistan (Amrao et al., 2010; Ullah et al., 2014a; 2015). CLCuBuV-infected plants normally show upward or downward curling of leaf margins, swelling and darkening of veins, which frequently develop into cup-shaped leaf-like out-growths called “enations”. Infected leaves become thickened and more brittle than those from healthy plants. Severely infected leaves can show rolling and a reduction in size, with spirally twisted leaf petioles, branches and the main stem (Akhtar et al., 2013; Siddique et al., 2014). Severely infected plants show a significant adverse effect on lint yield, agronomic and fibre quality traits of cotton, which ultimately affect yarn quality parameters (Akhtar et al., 2009). Management measures for CLCuD may include the control of vector (whitefly). The whitefly may develop pesticide resistance (Ahmad et al., 2003) and the intense application of pesticides may have deleterious effects on the environment (Pico et al., 1998; Palumbo et al., 2001). Hence, the development and use of resistant cotton varieties, is the only feasible, effective, economical and environment friendly approach against CLCuD.
Cotton, being an important cash crop, is also vulnerable to be attacked by a large number of pest insects, throughout its growth period causing serious yield and economic losses (Akram et al., 2013; Ullah et al., 2014b). Growing of healthy cotton plants, against pest ravages is a lifeline for local and foreign textile industries (Sarwar, 2013a; b). To control these pests large amount of broad-spectrum chemical pesticides are used. Most of these pesticides are found to be neurotoxic to humans and carcinogenic in nature. Many of these chemicals persist in the environment causing pollution to land and water resources (Damalas and Eleftherohorinos, 2011). This practice also has resulted in the development of pest resistance against most of these insecticides after repeated uses over the years (Ishtiaq and Saleem, 2011; McCaffery, 1998). To solve these problems Bt-cotton (Genetically Modified, Insect resistant) has been developed and adopted by farmers in several parts of the world. Commercial release of Bt-cotton with genes from the soil-born bacterium Bacillus thuringiensis has been one of the most successful developments in this area (Ullah et al., 2014b). In Pakistan, exotic Bt-cotton varieties entered through non-formal channels and their cultivation was first reported in the year 2000 in Sindh province with the cultivation of unauthorized Australian cotton variety Bollgard I - MON 531 event (USDA GAIN, 2010; James, 2010). Later, breeders from public and private sector developed Bt varieties in local genetic back ground through backcrossing using exotic Bt-cotton containing Cry1Ac gene (MON 531 event). Like other parts of the world, adoption of Bt technology in Pakistan is unprecedented. In 2010, as a trial experiment around 600,000 farmers planted Bt-cotton on more than 60% area (USDA GAIN, 2010; James, 2010). After getting satisfactory results from Bt-cotton varieties, and continuous requests from the farmers and the scientific community, the Government of Punjab province approved Bt-cotton for general release and cultivation (Arshad et al., 2007).
The exact status regarding the response of Bt-cotton genotypes to CLCuD is not clear which is the dire need of present situation. So, there is an urgent need to screen Bt-cotton genotypes against CLCuD. But, work done in this regard is still limited and needs more extensive research. Thus, the present study was under taken to find out the exact degree of resistance/susceptibility of available Bt-cotton genotypes under high inoculums pressure in the field and using graft inoculation in glasshouse by visual symptom scoring assessments.
Materials and Methods
Field testing
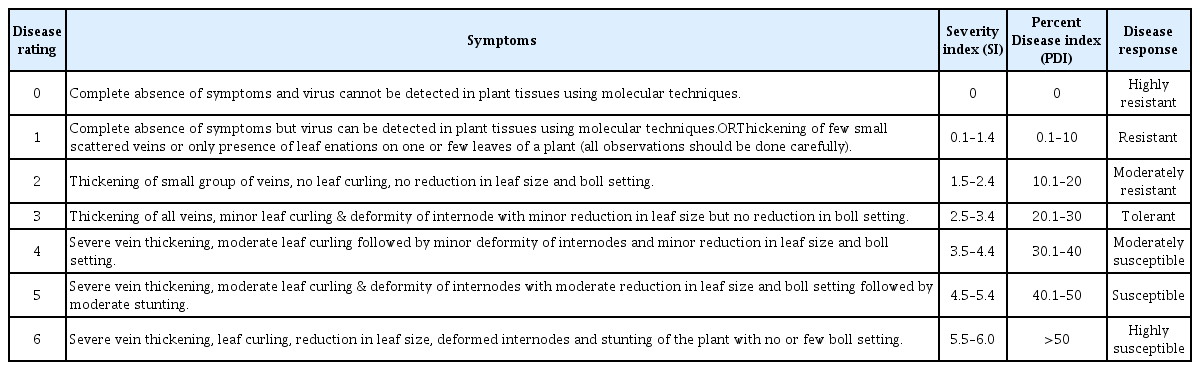
The response of 75 Bt-cotton genotypes was evaluated under natural field conditions from 2011 to 2013 in National coordinated varietal trial and Provincial coordinated cotton trial at the Nuclear Institute for Agriculture and Biology (NIAB), Faisalabad, Pakistan. Each genotype was planted in triplicates following Randomized Complete Block Design (RCBD) and conventional agronomic practices were employed but no pesticides were applied against whitefly vector to maximize inoculum pressure throughout the experiment. Data for CLCuD were recorded following the rating system described in Table 1 to calculate severity index (SI), percent disease index (PDI) and response of genotypes as suggested by Akhtar et al. (2010).
Graft inoculation evaluation
Five six-week old plants of each test genotype were inoculated by graft inoculation with CLCuBuV according to the method described by Akhtar et. al. (2013). For graft-inoculation, a sliced cut was made with the help of a scalpel near the tip of test plant. A fresh symptomatic branch infected with CLCuBuV was detached from glass-house maintained diseased plants and a similar cut was also made on this branch. Then cut surfaces were brought together and tied with parafilm. The symptomatic cut stem was then dipped in a test tube filled with distilled water. Distilled water was replaced daily and after seven days the tubes were removed (Akhtar et al., 2013). Experimental unit was observed daily and data were recorded on the percentage of successful grafts, percentage of disease transmission, latent period (average time required for first symptom appearance after grafting), and SI 90 days post inoculation (DPI) using the rating system described in Table 1.
Results and Discussion
CLCuD caused by CLCuBuV is presently a serious constraint in cotton production across Pakistan and north-western India (Ullah et al., 2014b). Limited work on screening of Bt-cotton against CLCuBuV has been conducted in Pakistan (Personal communication). Hence, the response of indigenous Bt-cotton genotypes to CLCuBuV is not clear. Present study reports a comprehensive evaluation of levels of resistance/ susceptibility to CLCuBuV in large number of Bt-cotton genotypes for the first time under high inoculum pressure in the field and using grafting in the glasshouse.
Results of field evaluation of cotton genotypes to CLCuBuV differed greatly from those of grafting experiments. The 75 Bt-cotton genotypes that were tested in the field from 2011 to 2013 were all infected systemically with CLCuBuV and showed different responses to infection by CLCuBuV based on symptomatology. None of the tested genotype was found to be disease free (highly resistant) during 2011–2013. The percent disease index (PDI) varied from 5.4 to 45.6% during 2011; 27.3 to 99.3% during 2012 and 37.2 to 98.6% during 2013 with SI values of 2.2 to 4.3 in 2011; 2.4 to 5.96 in 2012 and 3.3 to 5.9 in 2013, respectively (Table 2). During 2011, 3 genotypes were found resistant, 4 moderately resistant, 3 tolerant, 2 moderately susceptible and one susceptible; during 2012, 3 genotypes were found tolerant, 7 moderately susceptible, 5 susceptible and 38 highly susceptible; during 2013, one genotype was noted moderately susceptible and 51 were highly susceptible.
Whiteflies started to appear in low densities just after the emergence of seedlings and continued their build up during the whole growth period of cotton during 2011–2013. The first disease symptoms were initiated as thickening of small veins and curling of leaves on few young leaves of a small number of plants after 30–35 days of sowing during all three years. PDI and SI values increased with the passage of time depending upon the genetic makeup of genotypes. Plants infected at an early stage of growth exhibited severe symptoms, while mid season and late infections were milder and expressed reduced symptoms. Severe symptoms started with a minor vein thickening of young leaves which progressed to severe vein thickening, leaf curling, reduction in leaf size, deformed internodes and culminated in severe stunting of the plants with no or few bolls. The genotypes showing resistant to tolerant response during one year became moderately susceptible to highly susceptible in the next year. These results are in accordance with the reports of Akhtar et al. (2010) who performed the screening of large number of non-Bt-cotton genotypes belonging to G. hirsutum species against CLCuD under field and found the same trends.
Two genotypes, viz Sayban-201 and A-555 showed low level of disease during two testing years compared to other genotypes. Sayban-201 showed tolerant response during 2011 with PDI of 24.8% and SI value of 3.1 while showed moderately susceptible response during 2012 with PDI of 35.3% and SI value of 3.6. Genotype A-555 showed moderately susceptible response during two consecutive years (2012 and 2013) with PDI of 34.1% and 37.2% and SI values of 3.2 and 3.3 respectively (Table 2). Cauquil and Folin (1983) showed that CLCuV transmission efficiency is greater when there were 10 or more whitefly per plant. In present study we also observed the higher number of whiteflies (more than 10 per plant) throughout the experimental period which caused severe disease. The low level of disease exhibited by A-555 at field level may be due to the inability of whitefly to introduce a sufficient quantity of virus particles into the plants to cause a severe disease response as reported earlier in case of tomato against tomato leaf curl geminivirus and in case of non-Bt Gossypium specie against CLCuBuV (Akhtar et al., 2010; Tripathi and Varma 2002). Under field conditions, natural infection may not produce accurate results even if the studies are conducted in field with high vector populations (Sudha et al., 2013). An effective screening method is a critical component of an accurate disease assay for quantifying the levels of disease resistance among crop cultivars (Kim et al., 2014). Cotton genotypes evaluation against CLCuD by exposing them to high inoculums while planting in natural hot spots has been a principal method. One of the difficulties in selecting a virus resistant genotype under natural conditions is that the disease might occur with varying degrees of incidence as well as severity (Akhtar et al., 2010; 2011; Delatte et al., 2006). Such variable levels are thought to be due to the lack of a single factor or a combination of factors such as spatial and temporal variations in inoculum levels, environmental conditions, vector host preference, host resistance to vectors, age of plants, soil condition, etc. (Akhtar et al., 2010; Hoogstraten, 1992). To overcome these difficulties, field findings were further confirmed with artificial inoculation through graft inoculation.
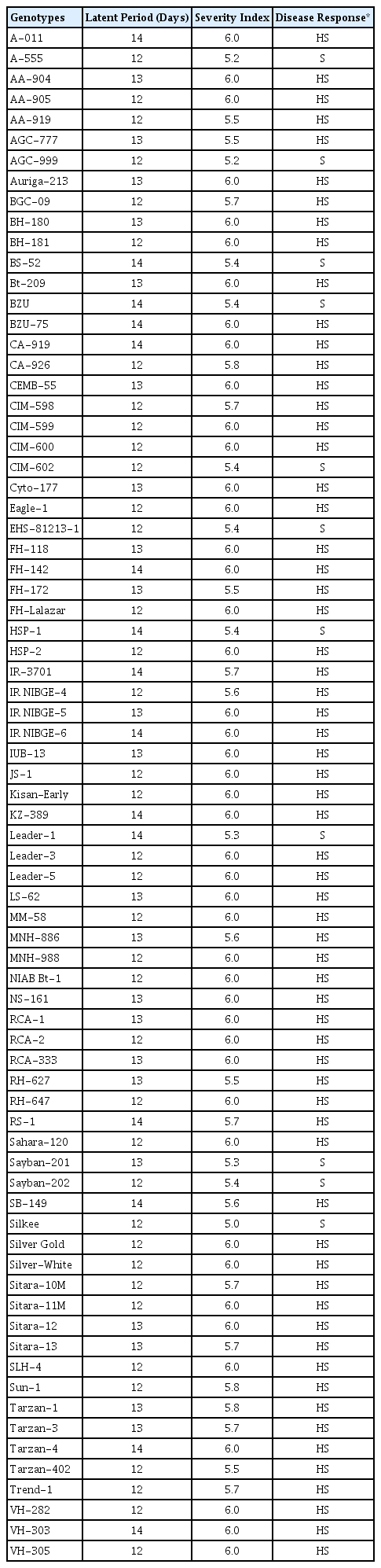
The evaluation results of 75 Bt-cotton genotypes against CLCuBuV through artificial inoculation (grafting) are presented in Table 3. Success of grafting and disease transmission was 100% in all tested genotypes. None of the graft inoculated plant was symptomless. All tested genotypes showed disease symptoms with SI values ranging between 5.0 and 6.0 and latent periods (LP) between 12 and 14 days. Of the 75 genotypes evaluated using graft inoculation, 11 were found susceptible with SI values of 5.0 to 5.4 while remaining 64 were highly susceptible with SI values of 5.5 to 6.0. Enation was observed in all genotypes belonging to both susceptible and highly susceptible categories. Inoculated plants of all tested genotypes exhibited severe disease symptoms like vein thickening, leaf curling, deformation of leaf petioles and shorting of internodes, etc., within 10 days after the appearance of initial symptoms. No reduction in SI values was observed until the end of the experiment i.e., 90 days after grafting. Akhtar et al. (2010) also reported that through grafting no source of resistance is present in non-Bt-cotton genotypes belonging to G. hirsutum species.
A successful plant breeding programs for disease resistance depends on the effective identification of sources of resistance and accuracy in resistance assessment technique (Akhtar et al., 2010; Pico et al., 1998). The current results obtained through natural transmission of virus by vector whitefly under natural field conditions and artificial inoculation through grafting under glasshouse conditions showed that the disease response of Bt-cotton genotypes varied according to the virus transmission methods employed. Field screening is a routine procedure and cannot be treated as a reliable procedure since it is seasonal and depends on epidemic conditions and may produce misleading results as has been evident from current findings. On the basis of present findings it can be concluded that graft inoculation is the most suitable, efficient and adequate method, and deemed practical in terms of screening. However, the field screening does not discriminate between these levels but can serve as a first step for further studies using graft inoculation.
Recent studies proved that Bt-cotton has no direct impact on the population dynamics of sucking insect pests (Ullah et al., 2014c). Akram et al. (2013) proved that Bt-cotton genotypes are more susceptible host for whiteflies and thrips than non-Bt-cotton genotypes. Population dynamics of sucking complex is found to be increased in Bt-cotton growing areas mainly due to reduced utilization of broad spectrum synthetic insecticides and low innate immunity of cultivars. This situation can create new problems in Pakistan (Khan et al., 2014). In Mali the introduction of Bt-cotton and the consequent reduction of insecticide treatments against key pests of cotton increased the incidence to 16SrII-C phytoplasma (Marzachiet et al., 2009). Recently Manzoor et al. (2014) reported a new isolate of Chickpea chlorotic dwarf virus (CpCDV) from cotton in Pakistan. This virus has been previously reported to be transmitted by leafhopper Orosius albicinctus Distant in other plant species (Akhtar et al., 2011), suggesting the presence of this leaf hopper spices in cotton. Orosius albicinctus is also recognized as the principal vector transmitting different kinds of phytoplasmas worldwide including in Pakistan (Akhtar et al., 2013). So, the suspected presence of O. albicinctus in cotton and increasing trend of Bt-cotton may create a problem to cotton which is already badly suffering from CLCuBuV.
Knowledge of resistance sources is useful in the selection of germplasm to incorporate in the breeding programs (Larsen and Porter, 2010). The management of CLCuD relies upon the resistance breeding programs, which involves identification of resistance sources and introgression of resistance genes into desired genotypes. The present research communication summarizes the results of our efforts to facilitate breeding of Bt-cotton for resistance to CLCuD caused by CLCuBuV. The results presented here clearly demonstrate that no sources of resistance to CLCuBuV are available among the tested Bt-cotton genotypes. By using the susceptible genotypes there are chances to increase CLCuD incidence and epidemic conditions for its development every year which may increase the possibilities of emergence of resistance breaking strains of virus, as previously reported (Akhtar et al., 2010a). Secondly, the increasing population dynamics of sucking complex in Bt-cotton growing areas due to the reduced utilization of broad spectrum synthetic insecticides and low innate immunity of cultivars can create new problems in Pakistan as mentioned above. So, a breeding program is needed to introgress the resistances against sucking insect pests and CLCuD from other resistant sources to agronomically suitable Bt-cotton genotypes. Information generated under the present study also provides a basis for further investigations concerned with epidemiology of the disease and alerts us to pay more attention to set up management strategies to prevent potential outbreaks of new pathogens from cotton.
Acknowledgements
The authors are thankful to Mr. Muhammad Tanvir Elahi (Research Associate), NIAB, Faisalabad for his useful assistance in data collections.