Rhizosphere Inhibition of Cucumber Fusarium Wilt by Different Surfactin- excreting Strains of Bacillus subtilis
Article information
Abstract
Bacillus subtilis B006 strain effectively suppresses the cucumber fusarium wilt caused by Fusarium oxysporum f. sp. cucumerinum (Foc). The population dynamics of Foc, strain B006 and its surfactin over-producing mutant B841 and surfactin-deficient mutant B1020, in the rhizosphere were determined under greenhouse conditions to elucidate the importance of the lipopeptides excreted by these strains in suppressing Foc. Results showed that B. subtilis strain B006 effectively suppressed the disease in natural soil by 42.9%, five weeks after transplanting, whereas B841 and B1020 suppressed the disease by only 22.6% and 7.1%, respectively. Quantitative PCR assays showed that effective colonization of strain B006 in the rhizosphere suppressed Foc propagation by more than 10 times both in nursery substrate and in field-infected soil. Reduction of Foc population at the cucumber stems in a range of 0.96 log10 ng/g to 2.39 log10 ng/g was attained at the third and the fifth weeks of B006 treatment in nursery substrate. In field-infected soil, all three treatments with B. subtilis suppressed Foc infection, indicated by the reduction of Foc population at a range of 2.91 log10 ng/g to 3.36 log10 ng/g at the stem base, one week after transplanting. This study reveals that the suppression of fusarium wilt disease is affected by the effective colonization of the surfactin-producing B. subtilis strain in the rhizosphere. These results improved our understanding of the biocontrol mechanism of the B. subtilis strain B006 in the natural soil and facilitate its application as biocontrol agent in the field.
Fusarium wilt disease (FWD) caused by Fusarium oxysporum f. sp. cucumerinum (Foc) occurs worldwide and can result in severe losses in agricultural yield (Jenkins, 1983; Martínez, 2003; Owen, 1955). Disease severity increases with planting time as demonstrated by the high disease incidence (>70%) in monoculture plots cultivated for over three years, resulting in great yield losses (25%–70%) (Owen, 1955, 1959; Vakalounakis, 1996). In the recent years, FWD occurred in the Jilin, Shandong, and Hubei Provinces in China (Jiang et al., 2012; Vakalounakis et al., 2004). The use of beneficial organisms to manage the disease is desired to avoid the harmful effects of chemical pesticides on human health. Among the identified beneficial organisms, Bacillus spp. is preferred because it produces endospores that are resistant to poor environmental conditions. Many strains of the Bacillus genus have been developed as biopesticides for the control of plant diseases (Copping et al., 2004; Fravel, 2005) through multiple approaches. These approaches include antibiosis, competition, rhizosphere colonization, cell wall degradation, and induced resistance (Heydari and Pessarakli, 2010). Among the 20 antibiotics (Stein, 2005) produced by Bacillus spp., the activities of the lipopeptides of the surfactin, iturin, and fengycin families have been extensively studied in vitro (Cho et al., 2003; Raaijmakers et al., 2010; Romero et al., 2007). Surfactin is a family of cyclopeptides (C12–C16) that show surfactant activity and bactericidal effects. Iturin (C14–C17) and fengycin (C14–C18) are cyclopeptides with antifungal activity (Ongena and Jacques, 2008; Raaijmakers et al., 2010). However, the function of these lipopeptides in the soil remains to be clarified.
Previous research disclosed that fengycin and iturin are involved in the inhibition of fungal growth (Asaka and Shoda, 1996; Liu et al., 2011) and disease suppression (Leclère et al., 2005) using mutants with deleted or overexpressed genes. The function of surfactin was also studied using mutants with deleted surfactin encoding genes (Bais et al., 2004). However, deletion of the surfactin synthase gene also destroys the ability of Bacillus to colonize on plant roots and may further affect the production of other lipopeptides, such as fengycin or iturin, in the rhizosphere. Thus, the use of mutants lacking the surfactin encoding gene is not a suitable method in clarifying the function of surfactin itself or its combined effects with other lipopeptides. Moreover, multiple function-change of Bacillus caused by gene deletion should be considered. This has been described by Xu et al. (2013) who found that bacillomycin D affects kin C gene expression and biofilm formation on root besides the inhibition of F. oxysporum growth.
The elucidation of this methodological issue may be resolved by the detection of lipopeptides from rhizosphere (Kinsella et al., 2009) or in plantlets (Henry et al., 2011; Nihorimbere et al., 2012). Through direct detection, surfactin has been proven to be the dominant lipopeptide in tomato plantlets (Henry et al., 2011; Nihorimbere et al., 2012), and acts as an elicitor that stimulates plant immune-related responses and suppresses Botrytis cinerea attack on tobacco leaves (Cawoy et al., 2014). Li et al. (2007) demonstrated that the biocontrol ability of B. subtilis mutant Bs-H74, which exhibits a threefold increase in surfactin production in nutrient broth, increases by 14.6% in suppressing rice blast. However, whether surfactin could suppress soil pathogenic fungi directly in rhizosphere has not been fully determined yet.
B. subtilis B006, which effectively suppresses cucumber fusarium wilt and chilli root rot, has been shown to produce surfactin and fengycin in nutrient broth and in the rhizosphere (Yang et al., 2012). To elucidate the importance of surfactin excreted by B. subtilis in its colonization and interaction with Foc in natural soil, we generated two genetically stable surfactin mutants, over-production mutant B841 and reduction mutant B1020, by nitrogen ion beam implantation from B. subtilis B006. Their activities in suppressing the cucumber FWD and Foc population in the rhizosphere were investigated in this study.
Materials and Methods
Three strains of B. subtilis and specificity of their genetic markers
The antagonistic B. subtilis strain B006 was obtained from our previous research (Yang et al., 2012). Mutant strains, B841 and B1020, were generated by nitrogen ion beam implantation from strain B006 (Jia et al., 2013). B841 has high surfactin-producing ability in nutrient broth, compared with strain B006, whereas B1020 has low surfactin-producing ability in nutrient broth. The strains were maintained on nutrient agar slants at 4°C.
The mutant sites of B841 and B1020 are not easy to identify. Therefore, the specificity of the primer pair SCAR-A/SCAR-S (SCAR-A 5′-ACCCCCGAAGCTGATTA-3′, SCAR-S 5′-TGGCCTTTTCTTAGTTGTC-3′), which is used to detect B006 in the rhizosphere (Geng et al., 2011; Yang et al., 2014), was first confirmed for the amplification of B841 and B1020. Sterile distilled water and strain B006 was used as negative and positive controls, respectively. Total DNA was extracted using the TIANamp bacterial DNA kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Real-time PCR was performed using the BioRad iQTM 5 Multicolor RT-PCR Detection System (Bio-Rad) with SYBR Premix DimerEraser (TaKaRa) containing SYBR Green I. Amplification was performed as previously described by Yang et al. (2014). Cycle threshold (Ct) values were collected using BIO-RAD iQ™ 5 software.
Inhibitive abilities of strain B006 and its mutants
Nihorimbere et al. (2012) found that organic acids, instead of sugars or other components in the root exudate, affect the total amounts and the constitution of the surfactin components. The cucumber root exudate (CRE) used in the present study was designed based on the description of Nihorimbere et al. (2012) and Kamilova et al. (2006) and is composed of the following: 0.5 g/l MgSO4·7H2O, 1.0 g/l K2HPO4, 0.5 g/l KCl, 1.0 g/l yeast extract, 1.2 mg/l Fe2(SO4)3, 0.4 mg/l MnSO4, 1.6 mg/l CuSO4 and 2 g/l (NH4)2SO4 (Nihorimbere et al., 2011) and 1.08 g/l glucose, 0.68 g/l fructose, 0.06 g/l maltose, 0.02 g/l ribose, 5.3 g/l citrate, 0.85 g/l succinate, 1.05 g/l malate, and 0.8 g/l casamino acids (Kamilova et al., 2006). The ability of the strain B006 and its mutants, B841 and B1020, to inhibit Foc growth was determined using the pair co-culturing method on 1% solid CRE medium at 30°C for 7 days. The radii from the colony center to the inhibitive zone (R1) and from the colony center to the normal growth margin (R2) were measured, and the inhibitive ratio (R1/R2) was calculated.
Detection of the lipopeptides excreted by strain B006 and its mutants
The lipopeptides produced by the three strains were also determined in CRE broth. The bacterial strains were inoculated in 50 ml sterilized CRE broth in triplicate and incubated at 30°C for 48 h with shaking at 180 rpm. Once the bacterial suspension reached an OD600 of 0.9, the lipopeptides were extracted and detected by high performance liquid chromatography-electrosprary ionization mass spectrometry (HPLC-ESI-MS) using the method described by Jia et al. (2013). After comparing the elution time and the m/z with the standard surfactin, the total amounts of surfactin and its each component were determined by calculating the relative peak areas in the chromatograms using MassLynx software. The fengycin was determined using the m/z with reference to a previous research by Yang et al. (2012), and its amount was also determined by calculating the relative peak areas in the chromatograms using MassLynx software.
Materials for planting in greenhouse tests
The cucumber cultivar “Zhongnong 6” with moderate resistance to FWD was procured from the Seed Corporation of the Chinese Academy of Agricultural Sciences (Beijing, China). The nursery substrate was obtained from the Langfang Seed and Seedling Corporation (Langfang City, Hebei Province). The nutrient components were similar to the substrate used by Yang et al. (2014), which contained 2% to 4% N P K, 30% firmuletes, and 42% organic materials. The pH was 5.5–6.5.
Biocontrol efficacy tests in the greenhouse
The biocontrol efficacies of strain B006 and its mutants, B841 and B1020, were tested using the nursery substrate and field soil in the greenhouse according to the scheme listed in Table 1. Different planting strategies and Foc inoculation methods were adopted in the two tests.
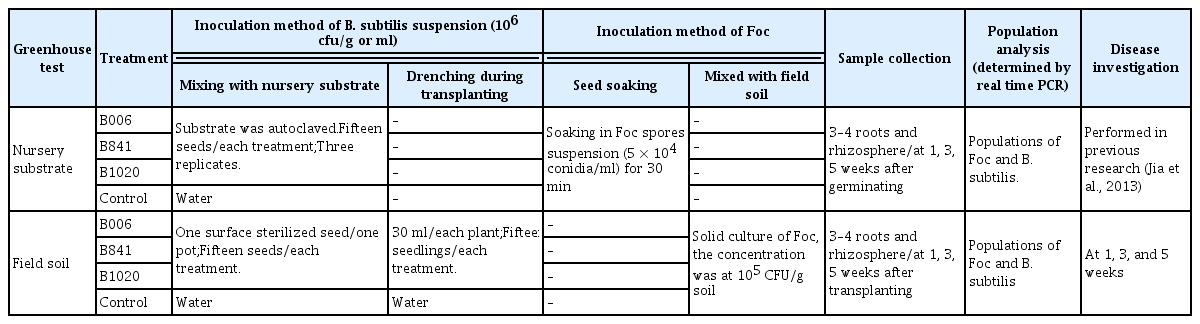
Experimental scheme of the biocontrol efficacy test for the three B. subtilis strains in the greenhouse
For the experiment using the nursery substrate, cucumber seeds were heated at 68°C for 3 h. Foc inoculation was performed by soaking the seeds in a Foc spore suspension (5×104 conidia/ml, 30 min). Ten seeds were randomly selected to determine the initial population of Foc on the seed surface using the serial dilution method. Fifteen seeds were sown in autoclaved nursery substrate that was pre-inoculated with fresh bacterial suspensions of the three strains at the concentration of 106 cfu/g. The initial concentration of each strain in the substrate was determined by the real-time PCR assay. Nursery substrate treated with autoclaved water served as control. Each treatment was tested in triplicate and was placed in the greenhouse at 25°C to 30°C. Three to four cucumber root samples and rhizosphere samples were randomly collected from each treatment at 1, 3, and 5 weeks after germination to determine the population of each strain. The Foc population in the stem and the rhizosphere were also assayed by real-time PCR.
The population dynamics of the B. subtilis strains and Foc in the rhizosphere were analyzed according to the method of Yang et al. (2014). The standard curve for the detection of strains B006, B841, and B1020 in the soil was first constructed using strain B006 based on the good specificity of the SCAR-A/SCAR-S primer pair determined in the initial real-time PCR analysis. The real-time PCR amplification of Foc was carried out with 10× diluted primers, FocF3 (5′-AAACGAGCCCGCTATTTGAG-3′) and FocR7 (5′-TATTTCCTCCACATTGCCATG-3′) (Lievens et al., 2007). The amplification program was similar to the method used by Yang et al. (2014) and was performed as follows: initial denaturation at 95°C for 1 min, followed by 45 cycles of denaturation at 94°C for 15 s, annealing at 62°C for 30 s, and elongation at 72°C for 30 s, and a final elongation at 72°C for 2 min. Melting curve conditions were: 81 cycles from 55.0°C to 95.0°C with a temperature rise of 0.5°C per 10 s. Real-time PCR standard curve was also constructed to determine Foc population in cucumber stem tissues. A total of 0.5 g of fresh healthy cucumber stem tissue was cut into pieces and mixed with 20 μl of Foc mycelial suspension at various concentrations (4 ng/g to 40 μg/g). The DNA was extracted using the FastDNA® SPIN Kit, and 2 μl of triplicate samples was analyzed by real-time PCR. Foc-free healthy tissue was used as a negative control.
The populations of the B. subtilis strains and Foc in all the collected samples were determined by real-time PCR and analyzed by iQTM 5 software using the same method described for the construction of corresponding standard curves.
For the experiment using field soil, the autoclaved nursery substrate was also inoculated with each tested strain at a final concentration of 106 cfu/g substrate. The cucumber seeds were first surface sterilized using 1% sodium hypochlorite and sown separately on the pre-inoculated nursery substrate. Autoclaved substrate treated with water served as control. The seed trays were placed in the greenhouse with the day/night temperature of 30°C/20°C. Cucumber seedlings with three to four leaves were transplanted to field soil pre-inoculated with solid cultures of Foc in the millet seed and rice hull medium (Burgess, 2008; Yang et al., 2014). The actual Foc population in soil was determined before transplanting. One seedling was planted per pot and then added with 30 ml of 106 cfu/ml bacterial suspension of each B. subtilis strain. This experiment was carried out at the greenhouse located in Langfang City, Hebei Province.
Cucumber root samples and rhizosphere samples were randomly collected from each treatment at 1, 3, and 5 weeks after transplanting and used to determine populations of the B. subtilis strains and Foc using the same method described above. The disease index was investigated at the fifth week according to the method described by Zhao and Wu (2001) and Yang et al. (2012). Control effect (%) = [(Disease index of the control − Disease index of treatment)/Disease index of the control] × 100%.
Data analysis
The data were statistically analyzed using SPSS 17.0 software, and significant difference was determined according to Duncan’s new multiple range test.
Results
Inhibitive abilities of strain B006 and its mutants
Strain B006, B841, and B1020 exhibited different colony morphologies on 1% CRE medium (Fig. 1A). Strains B841 and B1020 displayed slower growth in comparison with strain B006. The colony color changed from pale pink to milky white and was sticking on the CRE medium. Although changes occurred in the colony morphology, the inhibitory ratio (R1/R2) of B841 was still satisfactory at 0.37±0.03, which is comparable with strain B006 that had an inhibitory ratio of 0.40±0.02. The inhibitory ability of strain B1020 was lower, with a value of 0.52±0.02.
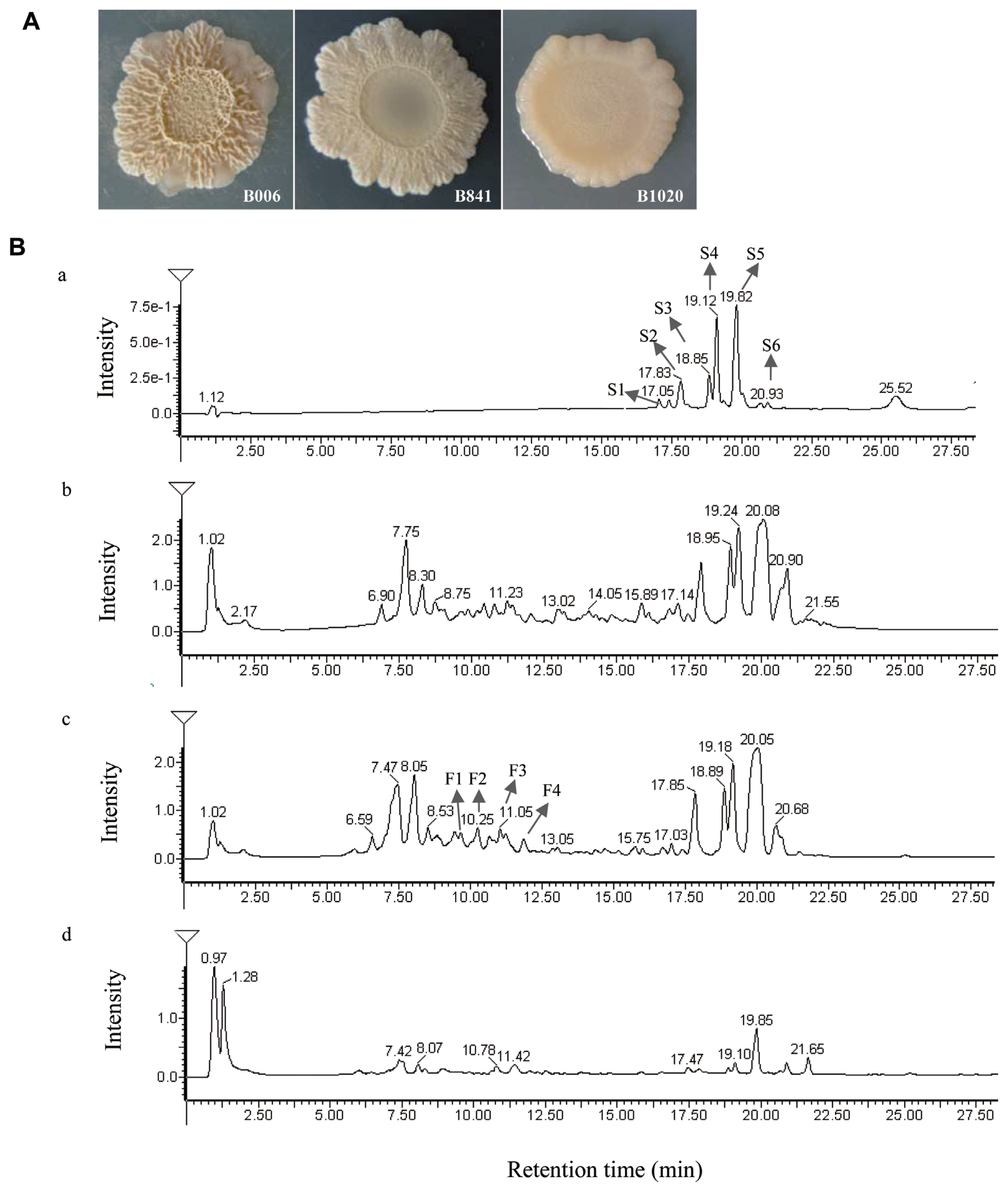
Morphologies of B. subtilis B006 and its surfactin mutants B841 and B1020 on CRE medium (A) and HPLC chromatograms of surfactin (a) and compounds extracted from cell-free supernatant of B. subtilis B006 (b) and its mutants B841 (c) and B1020 (d) in CRE broth (B). Retention time of surfactin was between 17.05 and 20.93 min.
Lipopeptides produced by strain B006 and its mutants
The retention time of standard surfactin was between 17.05 and 20.93 min (Fig. 1B, a), and the corresponding m/z was at 995 (C12), 1009 (C13), 1023 (C14), 1037 (C15), and 1051 (C16) (Supplementary Fig. S1A). In the extracts of B006, B841, and B1020, similar peaks appeared at the retention time between 17.14 and 21.55 min (Fig. 1B, b, c, d), and the corresponding m/z of peaks S1, S2, S3, S4, and S5 was the same with the standard surfactin components C12, C13, C14, C15, and C16, respectively. At the retention time between 9.03 and 12.25 min, the corresponding m/z of peak F1 and F2 (Fig. 1B, c) was 1436 and 1450, respectively, identical to the [M+H]+ of fengycin A C15 and C16 (Supplementary Fig. S1B), and the m/z of peak F3 and F4 (Fig. 1B, c) was 1478 and 1492, respectively, identical to the [M+H]+ of fengycin A C17 and fengycin B C16 (Supplementary Fig. S1B). The amounts of surfactin and fengycin produced by the three strains were compared after calculating the relative peak areas. The results showed that strain B841 produced more surfactin, whereas B1020 produced less in comparison with that produced by strain B006 (P < 0.05) (Fig. 2A). Strains B841 and B006 produced similar amount of fengycin, and strain B1020 produced the least, as indicated by the relative peak areas of 16051.7±424.16, 14810.5±892.87, and 5583.9±1215.37 (P<0.05) for B841, B006, and B1020, respectively. In addition to the total amount, the composition of surfactin changed. In comparison with the components produced by strain B006, the amounts of components C12, C13 and C16 produced by strain B841 were significantly higher (P<0.05), whereas the amounts of components C14 and C15 were at a similar level with strain B006 (Fig. 2B). The results indicated that the increased amount of components C12, C13, and C16 contributed to the increase in the total amount of surfactin produced by B841. The amount of each surfactin component produced by B1020 was significantly lower than that produced by other strains (Fig. 2B).

Comparison of the total surfactin amount (A) and each surfactin component amount (B) produced by strain B006 and its mutants B841 and B1020 in CRE broth. All data were analyzed by SPSS 17.0 software, and different letters above each column are significantly different according to Duncan’s new multiple range test at P < 0.05.
Standard curve of the real-time PCR assay of Bacillus and Foc
By using the primer pair SCAR-A/SCAR-S, a single 500 bp DNA band was amplified from the genomic DNA of mutants B841 and B1020, similar to that amplified from the wild type strain B006 (Supplementary Fig. S2A). Further amplification using the real-time PCR program proved that the SCAR primers were specific and suitable for the real-time PCR assays of the two mutants, indicated by the amplification of similar single curves with a melting temperature of 86°C (Supplementary Fig. S2B). Hence, a standard curve for real-time PCR of strain B006 and mutants B841 and B1020 was constructed using strain B006 as a model. Perfect amplification curves were obtained by real-time PCR for the detection of B. subtilis B006 and its mutants in the rhizosphere sample using the optimized program with 10× diluted primers. The standard curve for B006 concentration in the rhizosphere was linear with a range of 2.30×106 to 2.30×101 cfu/g, and the regression equation of linearity Y = 0.9863X − 1.1134 (r2=0.995) was obtained.
For the determination of Foc population, standard curves for Foc detection in the rhizosphere and in cucumber tissues were also constructed using logarithm-transformed values of DNA copies plotted against logarithm-transformed values of Foc concentration to avoid the effect of the variation on the DNA extraction from the rhizosphere or cucumber tissue samples. The standard curve for Foc concentration in the rhizosphere and in tissues was linear over the range of 4 ng/g to 40 μg/g, with regression equations of linearity Y = 0.513X + 1.4332 (r2 = 0.9851) and Y = 0.5856X + 0.6595 (r2 = 0.9845), respectively.
Suppression of Foc populations by B. subtilis colonization in the nursery substrate
The population dynamics of the B. subtilis strains and Foc were analyzed from the same set of samples. At the first week after germination, the population of B. subtilis in all three treatments decreased from 106 cfu/seed to 104 cfu/g substrate (Fig. 3A), whereas Foc number increased to 105 ng/g soil with no significant difference among all treatments (P ≤ 0.1) (Fig. 3B). At the third week, the B006 population increased to 3.14×104 cfu/g substrate, which is higher than that of strains B841 and B1020 (P ≤ 0.1) (Fig. 3A). At this point, the Foc population in the blank control was at 7.94×104 ng/g substrate, whereas the treated pots have lower Foc population, indicating that the treatments caused this decrease. Among the strains, B006 gave the best suppression, with a Foc population reduction of 1.0 log10 ng/g substrate, whereas B841 and B1020 had lesser suppression ability with Foc number reduction of 0.6 log10 ng/g soil (P ≤ 0.1) (Fig. 3B). At the fifth week, the populations of the three B. subtilis strains increased to 104 cfu/g substrate (Fig. 3A), compared with their populations at the third week. While the Foc population in all treatments also increased from the third week, indicating that Foc suppression by the treatment declined. However, the Foc number in samples treated with strain B006 was still the lowest among all the treatments (P ≤ 0.1) (Fig. 3B).
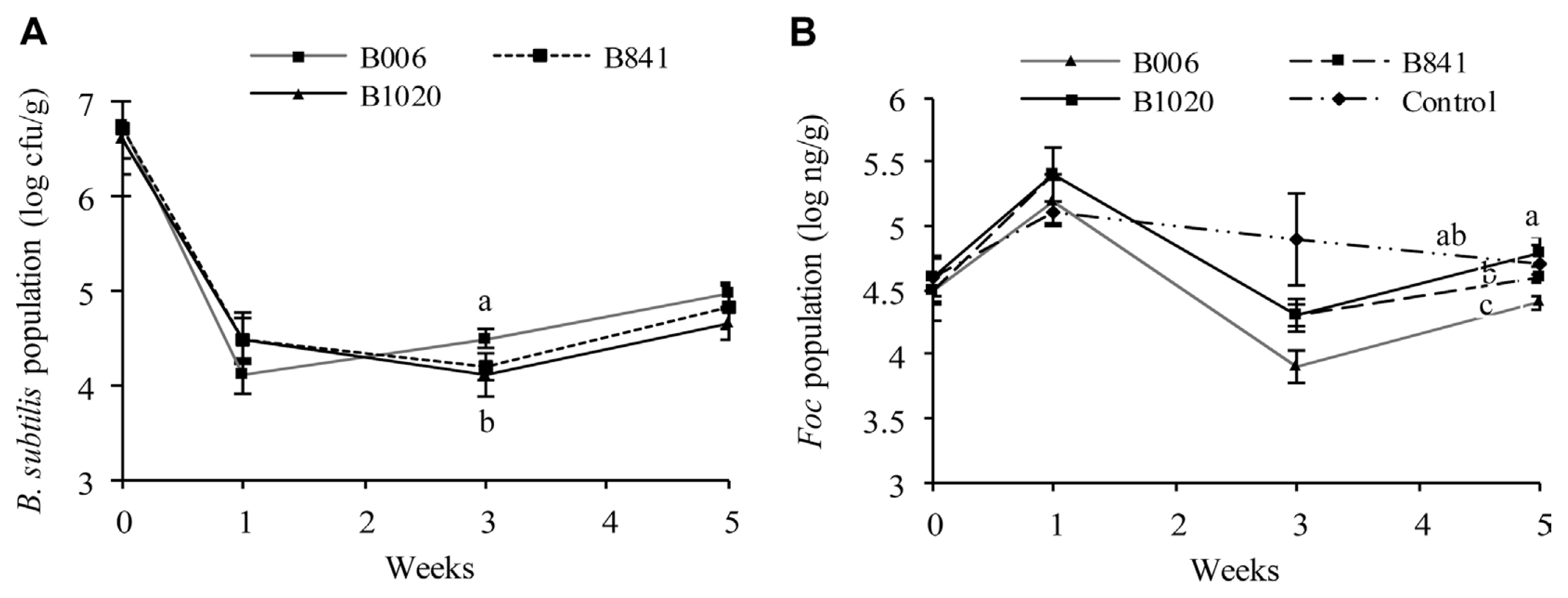
Populations of B. subtilis (A) and Foc (B) in cucumber rhizosphere determined by real-time PCR in sterile substrate with the addition of bacterial suspension of B. subtilis B006 and its mutants at 106 cfu/g. All data were analyzed by SPSS 17.0 software, and different letters above each column are significantly different according to Duncan’s new multiple range test at P ≤ 0.1.
The suppressing ability of the three strains was also verified by the real-time PCR assays of Foc in plant stems. At the first week, the Foc number in stems was similar without significant difference. At the third and fifth weeks, the Foc number in the cucumber stems of B. subtilis treatments reduced significantly (P ≤ 0.1) compared with the control, especially at the upper part of the stem (Table 2). Reductions of Foc population in the stems ranged from 0.96 log10 ng/g to 2.39 log10 ng/g and 0.31 log10 ng/g to 1.49 log10 ng/g at the third and the fifth weeks, respectively, treated by strain B006 and B841.
Suppression of Foc populations by B. subtilis colonization in the field soil
The initial population of B. subtilis strains B006, B841, and B1020 in the nursery substrate was at 2.3×106 cfu/g, 1.6×106 cfu/g, and 2.1×106 cfu/g, respectively. The population dynamics of the B. subtilis strains (Fig. 4A) and the Foc (Fig. 4B) were different among treatments. The B. subtilis populations in the rhizosphere of the treated field soil increased at one week after transplanting; The Foc populations in the treated soil were kept at a similar level among treatments. At the third week, the B. subtilis population in the B006 treatment reached the highest value of 8.71×104 cfu/g among all treatments, whereas the Foc population decreased to the lowest value of 3.39×103 ng/g (P ≤ 0.1). At the fifth week, the B. subtilis populations in the three treatments decreased, whereas the Foc populations increased. B006 also displayed the highest population of 5.37×104 cfu/g in rhizosphere (P ≤ 0.1). However, the Foc population in B006 treatment was still the lowest (1.82×105 ng/g) compared with the control (3×105 ng/g) and other treatments. Real-time PCR analysis of the Foc populations in cucumber stems showed that all of the three B. subtilis strains could suppress Foc infection, as demonstrated by the significant reduction of Foc populations at around 2.91 log10 ng/g to 3.36 log10 ng/g at the stem base, one week after transplanting. In addition, the application of B. subtilis suppressed the spread or multiplication of Foc at the fifth week, which exhibited population reduction in the third or fourth true leaves of the plants, at a range of 1.08 log10 ng/g to 1.55 log10 ng/g, in comparison with that of the control (Table 3).
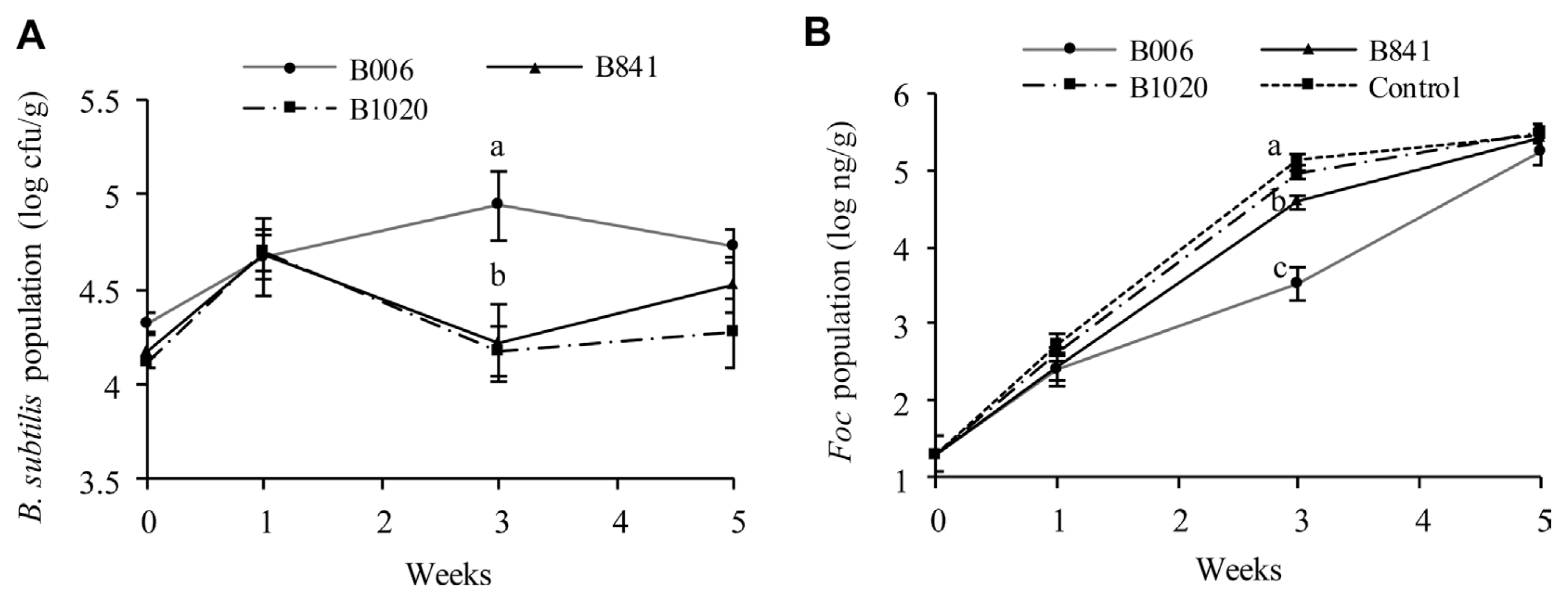
Populations of B. subtilis (A) and Foc (B) in cucumber rhizosphere in field soil. B. subtilis (B006, B841, and B1020) suspensions were inoculated in the nursery substrate at 106 cfu/g and were re-applied during transplanting. Data were analyzed by SPSS 17.0 software, and different letters above each column denote significant difference according to Duncan’s new multiple range test at P ≤ 0.1.
Control efficacy of B. subtilis B006 and its mutants in the field soil
Investigation of the disease index at the fifth week showed that B. subtilis strain B006 gave the best control performance in suppressing the development of fusarium wilt. The control efficacy was 42.9%, better than treatments with mutants B841 and B1020 (Table 4).
Discussion
Lipopeptides are considered to be the important characters for a bio-agent to be successful in field applications (Heydari and Pessarakli, 2010). Among lipopeptides, surfactin is not only involved in the colonization of antagonists in the rhizosphere (Cao et al., 2011; Fan et al., 2011; Hao et al., 2010), but also played important roles in disease suppression, in vitro or in bioassays (Keel et al., 1989; Thomashow and Weller, 1988). However, little is known whether lipopeptides are involved in the suppression of pathogen multiplication in the rhizosphere. Using surfactin overexpressed or deficient strain was an alternative way to determine the function of surfactin in the rhizosphere. In this study, we compared the wild surfactin-producing strain B006 (Guo et al., 2010; Yang et al., 2012) and its surfactin over-producing mutant B841 and surfactin-deficient mutant B1020 (Jia et al., 2013) for their abilities to colonize in the cucumber rhizosphere and suppress Foc multiplication. Our results showed that mutant B1020 had a similar colonization dynamics with mutant B841 in the rhizosphere (Figs. 3A and 4A), but was ineffective in disease suppression. The loss of the disease suppression capability was attributed to the population reduction in the rhizosphere and the deficient production of surfactin and fengycin, especially of the surfactin components C14 and C15. This result indicates that surfactin played an important role in the suppression of Foc multiplication and disease control. Henry et al. (2011) and Cawoy et al. (2014) found that adding 10 μM pure surfactin effectively suppressed disease development caused by Botrytis cinerea. Other researchers have detected enough surfactin from plantlets and the hydro-culturing tomato rhizosphere treated with bacterial strains of Bacillus spp. (Henry et al., 2011; Nihorimbere et al., 2012). Cawoy et al. (2014) also found wild strains with efficient surfactin-producing ability exhibited good control efficacy. However, the suppressing ability may be related to the components constitution. Henry et al. (2011) reported that homologues with the shortest lipid chains (C12 and C13) fail to induce any hydrogen peroxide release; by contrast, C14 and C15 surfactins trigger a significant response. Our analysis of the surfactin components demonstrated that mutant B841 produced more surfactin than strain B006, but this increase did not result in improvement of the suppressing ability (Tables 2 and 3) and biocontrol efficacy (Table 4). Further analysis of the surfactin components revealed that the increase in total surfactin production by the B841 strain was mainly caused by the increase of components C12, C13, and C16 (Fig. 2B). Therefore, the ineffective improvement of the suppressive ability of mutant B841 was ascribed to the increase of components C12, C13, and C16. This result suggests that the total amount of surfactin was not always positively related to Bacillus colonization and disease suppression in the rhizosphere. The structure and concentration of the surfactin components should be examined as well.
Our previous research showed that good colonization of strain B006 results in effective suppression of Foc in the rhizosphere and performs good disease control efficacy (Yang et al., 2014). In this study, B. subtilis strain B006 treatment also suppressed Foc spread and multiplication in stems (Tables 2 and 3), as well as the suppression of Foc multiplication in the rhizosphere. Because strain B006 is not an endophyte in cucumber and cannot directly suppress Foc in stems, induced systemic resistance (ISR) can be considered as the main mechanism of B006 for Foc suppression in stems. Bacillus spp. could produce many metabolites as elicitors of plant defense (Van Loon et al., 1998), such as lipopeptides (Jourdan et al., 2009; Ongena and Jacques, 2008), exopolysaccharides (Park et al., 2008), serine (Kilian et al., 2000), proteins (Li et al., 2006), preoxidase, chitinase and β-1,3-glucose (Bargabus et al., 2002), and some volatile compounds, such as 3-hydroxy-2-butanone (Ann et al., 2013), 2,3-butanediol and acetoin (Kloepper et al., 2004; Farag et al., 2006; Ryu et al., 2004), 3-pentanol and 2-butanone (Song and Ryu, 2013), and alcohols and ketones (Li et al., 2010). Previous reports suggest that surfactin is important in the suppression of plant diseases among these elicitors. Enough surfactin was detected in hydro-culturing tomato rhizosphere (Cawoy et al., 2014; Henry et al., 2011; Nihorimbere et al., 2012), whereas volatile emission was significantly reduced on tomato root exudate medium and only trace amounts of butanediol and acetoin could be detected in the gas phase of strains S499 and FZB42 (Cawoy et al., 2014), indicating that volatile compounds were not the major metabolites that elicit disease suppression. Among the metabolic substances listed above, B. subtilis strain B006 only produced surfactin and fengycin other than hydrolytic enzymes, and only enough surfactin was detected in cucumber rhizosphere (Yang et al., 2012). Surfactin may be the main compound that involved in the ISR and suppression of cucumber fusarium wilt. We are carrying out the direct drenching test to verify this hypothesis.
Gene-disrupting strains or mutants in other ways are often used to inspect the function of a gene (gene cluster) and its products. However, as a metabolite, surfactin synthesis is regulated by several global transcription genes, and these genes also regulate other gene’s expression. For example, AbrB gene encodes a repressor that controls the expression of genes involved in sporulation (Robertson et al., 1989) and the production of antibiotics (Park et al., 2012). Jung et al. (2012) reported that surfactin production can be improved by activating two competence-stimulating pheromones, ComX and the competence and sporulation factor, to stimulate the transcription of srfA operon. Compared with strain B006, the mutation in B841 resulted in increased surfactin production, whereas the mutation in B1020 resulted in the reduced production of both surfactin and fengycin, indicating that the mutation sites are different (Li et al., 1994). In addition to lipopeptide production, the morphology of mutant B1020 on CRE medium (Fig. 1A) also changed. The change in its physiological characteristics includes slow growth and reduced spore formation, as reported in our previous research (Jia et al., 2013). Therefore, the physiological characteristics should also be examined when a genetic mutant strain is used in an experiment.
Bacillus strains should be applied in an appropriate method to reach its best performance in suppressing disease. The wild type strain B006 performed the best suppression, both in rhizosphere (Figs. 3 and 4, Table 3; Jia et al., 2013) and in cucumber stems (Tables 2 and 3). However, if the seeds have been infected with Foc prior to the application of B006, the strain could not effectively suppress the infection of Foc (Table 2). Our previous research also demonstrated that adding B006 powder in nursery substrate can promote the effective colonization of B006 in cucumber rhizosphere before transplanting and suppress the development of FWD (Yang et al., 2014). All these results suggested that pre-application of B. subtilis strain B006 and the following induction of ISR were important for the effective suppression of Foc infection.
In conclusion, the results in this study suggest that the colonization of B. subtilis and suppression of Foc in the rhizosphere are important for fusarium wilt disease control. It was also found that the suppression is reduced with the reduction in the amount of total surfactin and its components, C14 and C15. In addition, increased surfactin production contributed by the increase of its components, C12, C13, and C16 did not enhance the suppression of Foc. These results improved our understanding of the biocontrol mechanisms of the B. subtilis strain B006 and will benefit its application in the field.
Acknowledgments
This study was supported by grants from the Programs of The National High Technology Research and Development of China (2011AA10A205), China Agriculture Research System (CARS-25-B-02) and “Five-Year” Plan of National Science and Technology Projects for the Rural Development in China (2012BAD19B0103).