A Genome-Wide Analysis of Antibiotic Producing Genes in Streptomyces globisporus SP6C4
Article information
Abstract
Soil is the major source of plant-associated microbes. Several fungal and bacterial species live within plant tissues. Actinomycetes are well known for producing a variety of antibiotics, and they contribute to improving plant health. In our previous report, Streptomyces globisporus SP6C4 colonized plant tissues and was able to move to other tissues from the initially colonized ones. This strain has excellent antifungal and antibacterial activities and provides a suppressive effect upon various plant diseases. Here, we report the genome-wide analysis of antibiotic producing genes in S. globisporus SP6C4. A total of 15 secondary metabolite biosynthetic gene clusters were predicted using antiSMASH. We used the CRISPR/Cas9 mutagenesis system, and each biosynthetic gene was predicted via protein basic local alignment search tool (BLAST) and rapid annotation using subsystems technology (RAST) server. Three gene clusters were shown to exhibit antifungal or antibacterial activity, viz. cluster 16 (lasso peptide), cluster 17 (thiopeptide-lantipeptide), and cluster 20 (lantipeptide). The results of the current study showed that SP6C4 has a variety of antimicrobial activities, and this strain is beneficial in agriculture.
The members of genus Streptomyces are a soil bacterium that produce various bioactive compounds (Bentley et al., 2002; Deshpande et al., 1988; Durán et al., 2018; Lee et al., 2020). Nearly 80% of the antibiotics used by humans are produced by Streptomyces (Durán et al., 2018; Harir et al., 2018; Hu et al., 2018). Many secondary metabolites produced by Streptomyces have antibacterial, antifungal, and anticancer properties (Genilloud, 2017; Gomes et al., 2017; Oskay, 2009). The genera Bacillus, Lactobacillus, Enterococcus, Streptococcus, and Lactococcus are known to produce ribosome-synthesized antibiotic peptides such as lantibiotics and bacteriocins (Cotter et al., 2005; Dischinger et al., 2009; Jabés et al., 2011). Nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) metabolites are synthesized by large enzymatic complexes that are mostly produced by the order Actinomycetales and Bacillus species (Ansari et al., 2004). Furthermore, the genus Streptomyces produce two-thirds of natural antibiotics, antitumor agents, and immune suppressors (Maiti et al., 2020).
The presence of Streptomyces spp. has been verified in the phyllosphere, rhizosphere, and endosphere of plants. The phyllosphere has recently drawn attention for being an important microbial habitat (Lindow and Brandl, 2003; Redford et al., 2010). In our previous study, Streptomyces globisporus strain S4-7 was well characterized as a responsible agent for the soil suppressiveness of Fusarium wilt of strawberry. This suggested the involvement of heat tolerance compounds and action in disease suppression (Cha et al., 2016). S. globisporus strain S4-7 produced a novel thiopeptide, conprimycin, which has several macrocyclic ring systems consistent with other thiopeptides (Cha et al., 2016). The bioactive conprimycin produced by S4-7 inhibits fungal cell wall biosynthesis and RNA polymerase activity (Cha et al., 2016). In addition, another antifungal lantipeptide gene (lanM) was identified and named grisin (Kim et al., 2019b).
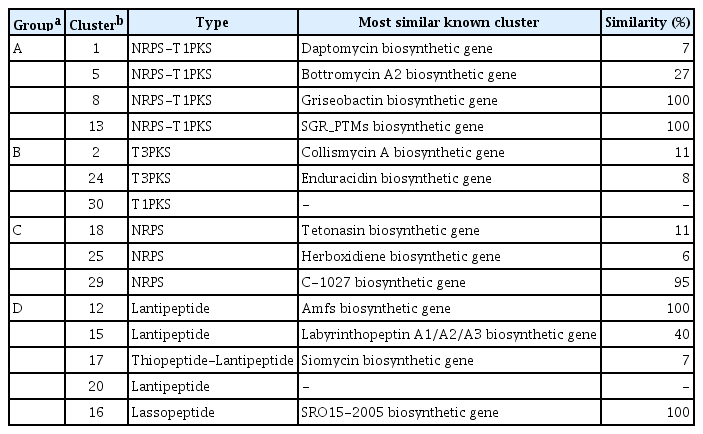
The genome of the bacterial strain used in this study has been reported by using shotgun genome sequencing (Cha et al., 2016) and PacBio genome sequencing reported by Kim et al. (2019a). We used antiSMASH (Medema et al., 2011) to predict the secondary metabolite gene clusters in this strain. The 15 predicted antibiotic-related genes were divided into groups of linked NRPS-PKS, PKS, ribosomal protein synthase-lantipeptide, and thiopeptide (Table 1, Fig. 1). Analysis of gene expression by real-time quantitative reverse transcription-polymerase chain reaction revealed that members of the antibiotic-related genes were reliably expressed, except for cluster 13 (Fig. 2A, Supplementary Table 1). To amplify the left arm (LA) and right arm (RA) regions for each target gene by CRISPR-Cas9 mutagenesis, SP6C4 genomic DNA (1 μg) and gene-specific primers were used (Supplementary Table 2). For each gene, (1) LA and RA regions were amplified using PCR under the following conditions: initial denaturation (at 98°C for 10 min) followed by 30 cycles of denaturation (at 98°C for 1 min), annealing (at 52–58°C for 30 s, respective primer tm), elongation (at 72°C for 30 s), and final extension (at 72°C for 5 min); (2) the LA and RA regions were linked by PCR (initial denaturation [at 98°C for 30 s], five cycles for denaturation [at 98°C for 8 s], annealing [at 50°C for 30 s], elongation [at 72°C for 30 s], and extension [at 72°C for 5 min]); and (3) the target homologous regions including LA and RA were amplified (the final PCR step was similar to [1] LA and RA region PCR conditions, which changed the annealing temperature to 60°C). PCR was performed using MJ Research PTC-200 Thermal Cycler (Bio-Rad, Hercules, CA, USA). The amplicons were loaded onto a 1% agarose gel and eluted in 30 μl of elution buffer (EB buffer) (Expin GelSV kit, GeneAll Biotechnology, Seoul, Korea). A-tailing was performed with 100 mM of deoxyadenosine triphosphate (dATP) (NEB, Ipswich, MA, USA) at 70°C for 30 min, the solution was purified with 5 volumes of GB buffer (Expin GelSV kit) and eluted in 20 μl of EB buffer. The purified molecule was ligated to a pGEM-T easy vector (Promega, Madison, WI, USA) with T4 ligase (Promega) by overnight incubation at 16°C and then transformed into Escherichia coli DH5α. Transformants were detected by blue/white colony selection on LB medium (Luria broth 30 g, agar 20 g/l) with ampicillin (100 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal, 40 μg/ml). Plasmids in white colonies were extracted using DokDo-Prep plasmid mini-prep kit (ELPIS-Biotech, Daejeon, Korea). Next, the vector and pCRISPomyces-gRNA (Cobb et al., 2015) were cut using XbaI (NEB, Ipswich, MA, USA), eluted in 30 μl of EB buffer, and a 3:1 ratio ligation was performed with 400,000 units (400 units/μl) of T4-ligase (NEB). The gRNA was cloned into pBHA (2,002 bp), the pCRISPomyces-2 was cut with BbsI (NEB), and the two fragments were ligated with T4 ligase and ligated again with the guide RNA (gRNA). The resulting plasmid was transformed into E. coli ET12567 containing pUZ8002 (Bierman et al., 1992), and transformants were recovered on LB agar containing kanamycin (30 μg/ml), chloramphenicol (25 μg/ ml), and apramycin (50 μg/ml) (Kieser et al., 2000). For seed cultures (E. coli ET12567/pUZ8002), single colonies were inoculated in LB broth containing the three antibiotics, then transferred to 10 ml of LB broth containing the antibiotics and incubated at 37°C for 9 h with shaking at 150 rpm. The cultured cells were first centrifuged (1,914 ×g for 15 min), then gently washed two times in 10 ml LB broth, centrifuged again, and thereafter suspended in 500 μl of LB broth. In a separate Eppendorf tube, 500 μl of 2× YT medium was heated to 50°C for 10 min and mixed with 10 μl of a spore stock of SP6C4 (109 cfu/ml) in 20% glycerol. The washed E. coli cells were added to the SP6C4 spore stock, mixed with a pipette, and spread on PDK agar (potato dextrose [Difco, Franklin Lakes, NJ, USA] 24 g, peptone 10 g, agar 20 g/l). After 16 h, apramycin (50 μg/ml) and nalidixic acid (25 μg/ml) were overlaid on the plates, and 7 days later, colonies were transferred two times onto PDK agar containing apramycin (50 μg/ml). Finally, each mutant was confirmed by PCR. All mutagenized strains and plasmids are shown in Table 2.

Schematic representation of deleted gene using CRISPR/Cas9 system. Left penal shows the distribution of gene functions and CRISPR/Cas9 targeted deletion sites. Group A was NRPS-PKS linked clusters, groups B and C were PKS and NRPS groups. Group D had lantipeptide and lassopeptide clusters. The cluster was characterized using antiSMASH version 4.0.0rc1 (https://antismash.secondarymetabolites.org/).
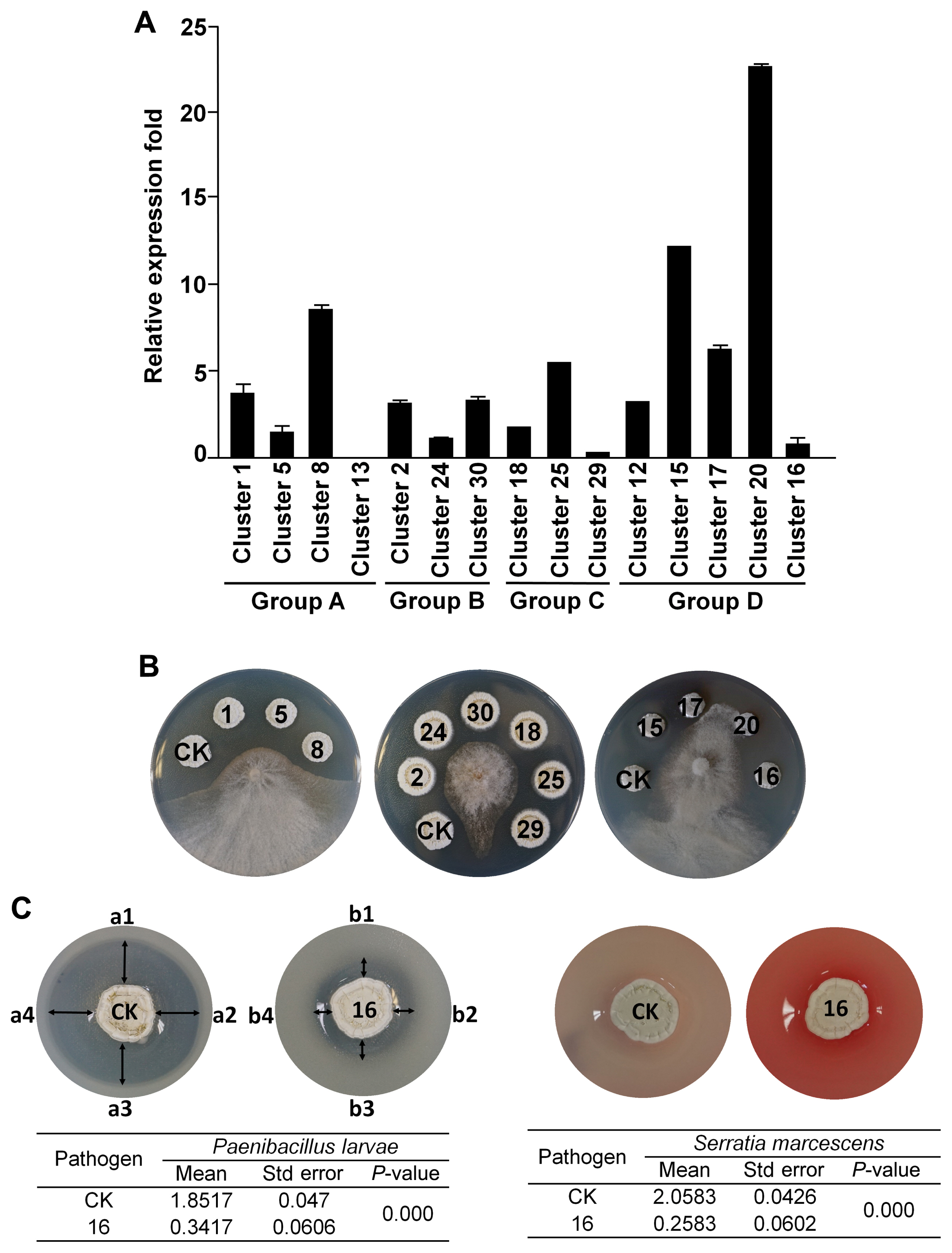
Antibiotic activity and related gene clusters in SP6C4. (A) Putative antibiotic gene clusters were predicted by antiSMASH, group A (NRPS-PKS), group B (NRPS), group C (PKS), group D (lantipeptide and lassopeptide). Relative gene expression levels were analyzed by qPCR. recA served as a housekeeping gene and primer information is presented in Supplementary Table 1. Bars represent standard error. (B) Antifungal activity of knockout mutants against B. cinerea. The mutants were created by the CRISPR/Cas9 system. The mutants (10 μl on a 0.8 cm filter disk) were incubated in PDK. After 5 days, an agar block of B. cinerea (0.4 cm diameter) was inoculated at the center of the plate and incubated at 28°C for 7 days (n = 5). CK represent the SP6C4 strain treatment and the numbers on the disc indicate each mutant based on the cluster number. (C) Paenibacillus larvae and Serratia marcescens served as entomopathogens. A two-layer method was employed to evaluate antibacterial activity. SP6C4 (106 cfu/ml, 10 μl) was inoculated on PDK medium. The entomopathogens were mixed into 25 ml of pre-cooled 0.2% PDK and overlaid onto the probiotic plates (final entomopathogen concentration was 105 cfu/ml). The overlaid plates were incubated at 28°C for 5 days and the size of the inhibition zone was measured (CK, a1 to a4; 16 cluster deletion mutants, b1 to b4). Entomopathogen inhibition test data were analyzed using one-way ANOVA, P-value less than 0.05 indicates that the size of the inhibition zones from CK and 16 was significantly different.
Except for the 14-cluster mutant strain, all other mutant strains showed no difference in growth and sporulation from the wild-type strains (data not shown). Antifungal Streptomyces mutant lines were confirmed using a plate assay. First, a single colony was streaked on PDK medium and cultured at 28°C for 5 days. Then the bacterial spore stock was collected with a sterilized cotton ball and filtered using a 10-ml syringe. Spore colony forming units were counted on PDK agar medium and 10 μl of samples were dropped onto 8 mm diameter paper disks on PDK agar. After sporulation (5 days post inoculation), 4 mm diameter plugs of actively growing mycelia of Botrytis cinerea were placed in the center of the plates. Fungal inhibition was evaluated after 7 days. This screening was repeated with 5 plates in each antiSMASH group and activity was defined as −, no inhibition; + (low inhibition), 0.1–0.5 cm; ++ (medium inhibition), 0.5–1 cm; +++ (strong inhibition), 1–1.5 cm; ++++ (maximum inhibition), >1.5 cm. Only the mutants in lantipeptide clusters 17 (tsrD, Conprimycin) and 20 (lanM, Grisin) were significantly less inhibitory to the gray mold pathogen (Fig. 2B). These results indicate that lantipeptide genes in the microbe play a major role in antifungal activity.
A two-layer method was used to evaluate the antibacterial activity against entomopathogenic bacteria, Paenibacillus larvae, and Serratia marcescens. Streptomyces strains were suspended to give 106 cfu/ml, and thereafter 10 μl was inoculated onto a 0.8 cm filter disk on PDK agar. P. larvae and S. marcescens were cultured in PDK broth for 3 days at 28°C, followed by mixing of 5 ml culture of each pathogen into 25 ml of pre-cooled 0.2% PDK agar and overlaying them onto plates. The plates were dried for 30 min, incubated at 28°C for 5 days, and antibacterial activity was indicated by the size of the inhibition zone around the disk. The entomopathogen growth inhibition assay was repeated five times as replications and analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (Tukey’s honestly significant difference) (P = 0.05) for mean separation. All graphs were visualized using ggplots version 3.0.1, and ggplot2 version 2.1.0 in the R software package. A mutant in cluster 16, encoding a lassopeptide, was less active against entomopathogenic bacteria (Fig. 2C). These findings support the hypothesis that the strain SP6C4 carries an arsenal of antibiotics needed to provide dual protection to plants and pollinators (Kim et al., 2019a).
The bacterial isolate in the rhizosphere has antifungal activity against strawberry gray mold fungal pathogens (Cha et al., 2016; Kim et al., 2019a). Studies on Streptomyces functional genes have rarely been conducted before using the CRISPR-Cas9 system. In this study, we used the CRISPR-Cas9 system (which is faster than homologous mutagenesis) to target mutagenesis, which involves the investigation of functional genes at a genome-wide scale. Moreover, our findings improved our understanding of the SP6C4 strain.
Notes
Conflicts of Interest
Youn-Sig Kwak serves as Editor-in-Chief for the Plant Pathology Journal, but has no role in the decision to publish this article. The remaining author has declared no conflicts of interest.
Acknowledgments
This research was supported by an agenda research program funded by the Rural Development Administration (PJ15871) and the Next-Generation BioGreen 21 Program (PJ013250).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).