Identification of strA-strB Genes in Streptomycin-Resistant Pseudomonas syringae pv. actinidiae Biovar 2 Strains Isolated in Korea
Article information
Abstract
Bacterial canker is a devastating disease of kiwifruit caused by the bacterium Pseudomonas syringe pv. actinidiae. Canker disease of kiwifruit in Korea has been controlled using streptomycin for more than two decades. Four streptomycin-resistant strains, belonging to biovar 2, which are found only in Korea, were collected between 2013 and 2014 from different orchards located in Jeju, Korea. The genetic background for streptomycin resistance among P. syringe pv. actinidiae strains were determined by examining the presence of strA-strB or aadA, which are genes frequently found in streptomycin-resistant bacteria, and a point mutation at codon 43 in the rpsL gene. All four streptomycin-resistant strains of P. syringe pv. actinidiae investigated in this study contained strA-strB as a resistant determinant. The presence of the aadA gene and a mutation in codon 43 of the rpsL gene was not identified.
Bacterial canker caused by Pseudomonas syringe pv. actinidiae (Psa) is a destructive disease of kiwifruit plants that has resulted in severe economic losses worldwide (Vanneste, 2017). Currently, the Psa population is divided into five subgroups called biovar 1, 2, 3, 5, and 6 (Sawada et al., 2016). However, evidence for the existence of a significant number of different populations within Psa has recently been reported (Mazzaglia et al., 2021). The strains belonging to biovar 1 were initially isolated from Japan and Italy in 1984 and 1992, respectively (ScoRtichini, 1994; Takikawa et al., 1989). Biovar 2 strains have been isolated only in Korea and cause severe economic losses on both green (Actinidia deliciosa) and yellow (A. chinensis) fleshed kiwifruit (Koh et al., 1994, 2010). The third population, biovar 3, which was first isolated in Italy in 2008 (Ferrante and Scortichini, 2010), has recently been responsible for a worldwide Psa pandemic (Balestra et al., 2013; Chapman et al., 2012; McCann et al., 2017). The biovar 5 and 6 strains were reported to occur in a limited region of Japan (Sawada et al., 2014, 2016). In Korea, biovar 2 or biovar 3 strains have been isolated from orchards infected with Psa (Lee et al., 2017).
Bacterial disease in plants has been controlled using chemical treatment methods and non-chemical strategies, such as cultural control practices, orchard management, activating the plant immune system, and breeding resistant varieties (Cameron and Sarojini, 2014). One of the chemical methods used to control bacterial pathogens in the field is spraying or a trunk injection of streptomycin (Koh et al., 2017). Streptomycin is an aminoglycoside antibiotic that inhibits protein synthesis in prokaryotes and has been used for agricultural purposes since the late 1950s (Sundin and Wang, 2018). The use of streptomycin for the control of bacterial pathogens on crops is legal in Asian countries, New Zealand, Israel, Canada, Mexico, and the United States, but its use in plant agriculture is prohibited in the European Union (Cameron and Sarojini, 2014). Streptomycin has been used since the 1990s to control bacterial canker of kiwifruit in Korea, the only country where the biovar 2 strain is isolated (Mazzaglia et al., 2012). Therefore, the appearance of the streptomycin resistance trait in biovar 2 means that it is the result of adaptation that occurred in Korea.
Most known streptomycin resistance determinants in bacteria encode enzymes that modify streptomycin by phosphorylation or adenylation. The strA, strB, and aadA genes are the three most widely distributed streptomycin resistance determinants (Sundin and Wang, 2018). Two phosphotransferase genes, strA and strB, which encode aminoglycoside-3″-phosphotransferase and aminoglycoside-6-phosphotransferase, respectively, were often linked to form a tandem gene pair, strA-strB. This gene pair is distributed worldwide among diverse gram-negative bacteria isolated from plants, animals, humans, and diverse environment samples (Petrova et al., 2008; Sundin and Bender, 1996; Tancos et al., 2016; van Overbeek et al., 2002). Additionally, the gene encoding an aminoglycoside adenylyltransferase, aadA, has been associated with streptomycin resistance in many clinical and phytopathogenic bacteria (Clark et al., 1999; Sundin and Wang, 2018; Xu et al., 2013).
Another resistance mechanism involves a point mutation at the target site of streptomycin. Streptomycin binds to the S12 protein, which is encoded by the rpsL gene, of the 30S ribosomal subunit, thereby inhibiting protein synthesis. A change in nucleotides at codon 43 of the rpsL gene from AAA, which encodes lysine in streptomycin-sensitive E. amylovora strains, to AGA (arginine), AAT/C (asparagine), or ACA (threonine) conferred a resistance phenotype to the antibiotic (Cameron and Sarojini, 2014; Chiou and Jones, 1995; Sundin and Wang, 2018).
The objective of this study was to identify the mechanism of streptomycin resistance in Psa biovar 2 strains isolated in Korea. Two possibilities for the resistance mechanism, enzymatic inactivation of the antibiotic and target site modification, were investigated.
The Psa strains used in this work were isolated between 2013 and 2014 from symptomatic kiwifruit plants in orchards located in Jeju, Korea (Table 1). The samples were surface sterilized using sodium hypochlorite, rinsed with sterile water, cut into small sections, and incubated in sterile water for 10 min with agitation. The suspensions were spread over peptone sucrose medium (PS; 20 g peptone, 20 g sucrose, and 15 g agar per liter), and the plates were incubated at 28°C for 24–48 h. The bacterial colonies were picked and transferred to PS agar to verify the pathogen. Identification of Psa and determination of biovars were conducted, as described previously (Lee et al., 2017). The strains identified as Psa were assessed for streptomycin resistance by streaking onto PS agar supplemented with 100 μg/ml of streptomycin. Streptomycin resistance was evaluated by observing the colonies on streptomycin-amended plates. Four streptomycin-resistant Psa strains were finally selected from different orchards, all of which belonged to biovar 2. Streptomycin-susceptible Psa strain JJ18 was included as a negative control.
The presence of strA, strB, and aadA genes in the four streptomycin-resistant strains was investigated by PCR using the primers developed by van Overbeek et al. (2002). Two primer pairs, aph(3″)-F/R and aph(6)-F/R, targeting two aminoglycoside phosphotransferase genes, were used for PCR amplification. A primer set, strA-F and strB-R, designed from strA and strB, respectively, were used to determine the existence of a linked strA-strB structure (Palmer et al., 1997). The aadA gene was searched using the primer set, ant(3″)-F/R. A pair of primers, rpsL212-F/R (Russo et al., 2008), was used to amplify the internal fragment of rpsL from the Psa strains. Table 2 lists the primer sequences and annealing temperatures for PCR. A PCR reaction mixture, 50 μl in total volume, consisted of 10 mM Tris (pH 8.0), 2.5 mM MgCl2, 0.2 mM of each dNTP, 2.0 U of Taq polymerase (Bioneer, Daejeon, Korea), 10 mM of forward and reverse primers, and 20 ng of template DNA. The PCR reactions were performed in a Thermal Cycler Dice (Takara Bio Inc., Shiga, Japan) under the following temperature conditions: 95°C for 5 min, 30 cycles of 95°C for 30 s, 30 s at each annealing temperature, and 72°C for 30 s, followed by a final extension at 72°C for 7 min. The PCR products were electrophoresed and visualized on a 1.2% agarose gel stained with ethidium bromide.
Two primer pairs, aph(3″)-F/R and aph(6)-F/R, amplified 515 bp and 560 bp of the internal fragments of the strA and strB genes, respectively, from streptomycin-resistant Psa strains and Escherichia coli DNRP545. On the other hand, no amplification occurred in the streptomycin-sensitive strain (Fig. 1A and B). In addition, the 1,571 bp amplicon was produced by PCR in streptomycin-resistant Psa using the forward primer designed from strA and the reverse primer from strB, indicating that the two genes are linked in tandem to form a gene complex (Fig. 1C). No amplification products were obtained from streptomycin-resistant Psa strains when the primers specific for aadA were used for PCR. Only E. coli DMRP5410 used as a positive control produced the expected 295 bp amplicon (Fig. 1D). Hence, the mechanism of resistance in Psa is due to the presence of the strA-strB gene cluster (Table 1).
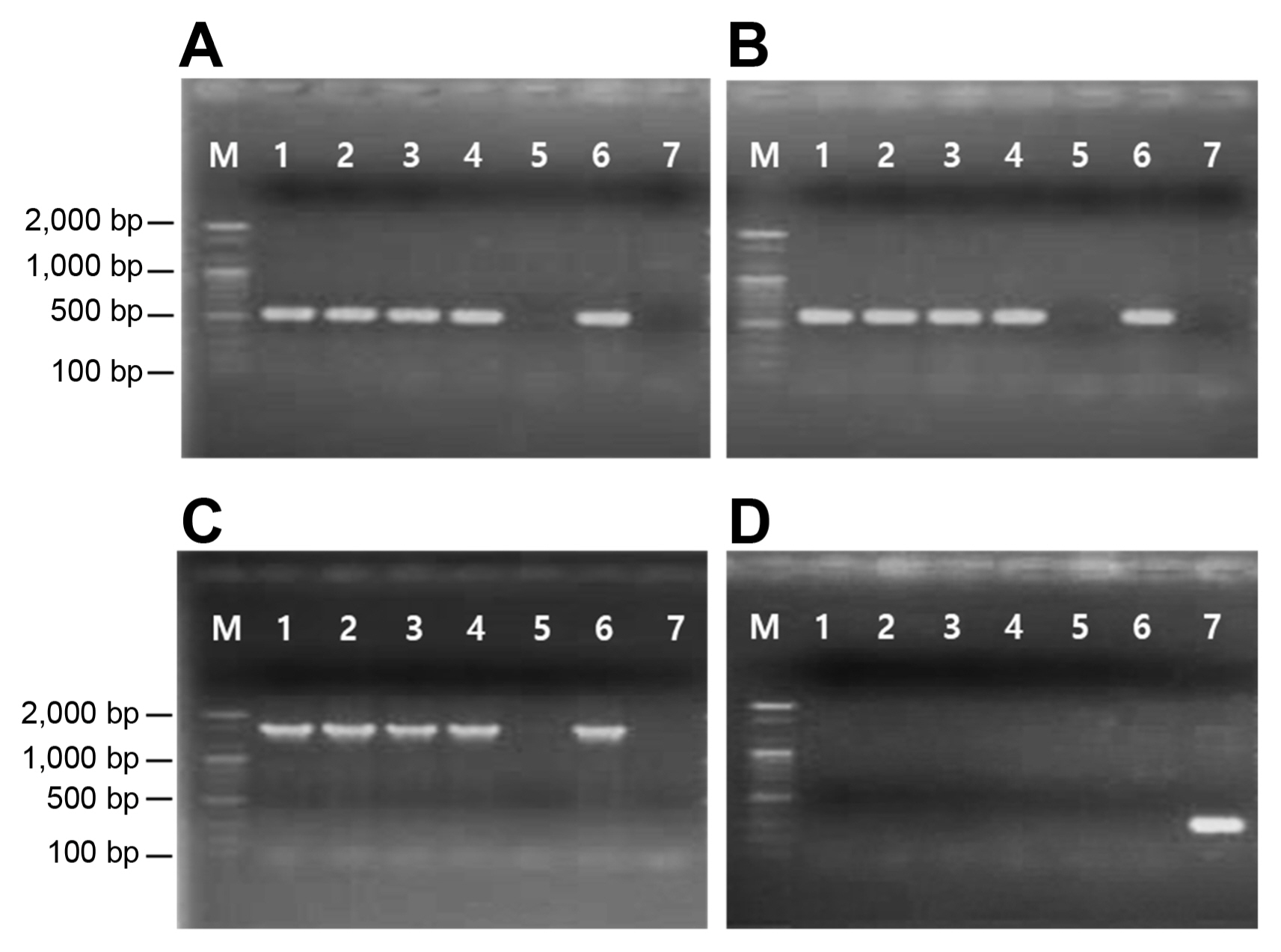
PCR detection of strA (A), strB (B), strA-strB (C), and aadA (D) genes. Lane M, size markers (100 bp ladder); lanes 1–5, strains of Pseudomonas syringae pv. actinidia: KSH1351 (lane 1), OYJ1351 (lane 2), KTS1431 (lane 3), KBD14311 (lane 4), and JJ18 (lane 5); lane 6–7, strains of Escherichia coli: DNRP545 (lane 6) and DMRP5410 (lane 7).
Sequence analysis of the rpsL gene in Psa was conducted to compare the nucleotide sequences of the gene of the streptomycin-resistant strains with those of the sensitive strain. After amplification, the PCR products of the rpsL gene were purified using an AccuPrep PCR Purification Kit (Bioneer). Nucleotide sequencing was performed in both the forward and reverse directions at SolGent Co. (Daejeon, Korea). A comparison of the nucleotide sequences of the PCR products amplified with rpsL212-F/R primers showed that all resistant strains had identical sequences to that of the sensitive one (data not shown). The four streptomycin-resistant strains had identical sequences to the susceptible strain JJ18 at a codon 43 of the rpsL gene coding for the amino acid, lysine (AAA). This indicates that streptomycin resistance in Psa is not due to a mutation in rpsL.
Of the 120 Psa strains isolated from Korea in 1998, there was no streptomycin-resistant strain. On the other hand, four P. marginalis strains isolated at the same place showed resistance to streptomycin, and three of them had the strA-strB gene pair as the resistant determinant. The resistance of one strain was attributed to a mutation in the target gene, rpsL, encoding a ribosomal protein S12 (Han et al., 2003). Hence, the strA-strB gene pair in the epiphytic bacterium, such as P. marginalis, may have migrated to Psa via horizontal gene transfer. No streptomycin-resistant strain was observed among the 21 Psa strains isolated in Korea in 1999 (Lee et al., 2005). In contrast, the streptomycin-resistant Psa strains isolated in Japan had the strA-strB genes or were caused by a mutation in the rpsL gene (Han et al., 2003).
The use of streptomycin in plant agriculture has resulted in the development of streptomycin resistance in pathogens worldwide (Förster et al., 2015; Sundin and Wang, 2018). Many studies of streptomycin resistance in phytopathogenic bacteria had been conducted with E. amylovora. The mechanism of streptomycin resistance found mainly in E. amylovora differed according to the regions where the bacteria had been isolated. Streptomycin resistance through a mutation in the rpsL gene was described in the isolates from New Zealand, Mexico, and several states in the United States, including California, Oregon, Washington, Idaho, Michigan, and Utah (Chiou and Jones, 1995; Nischwitz and Dhiman, 2013; Ponce de León-Door et al., 2013). Meanwhile, the strA-strB gene pair was the most common determinant of resistance in Michigan and New York (McGhee et al., 2011; Russo et al., 2008; Tancos et al., 2016). On the other hand, the genes associated with streptomycin resistance in some E. amylovora and Clavibacter michiganensis have not been identified, suggesting the existence of a new resistance mechanism (Lyu et al., 2019; Ponce de León-Door et al., 2013).
Our results indicate that streptomycin resistance of Psa biovar 2 strains isolated in Korea is due to the strA-strB genes. To understand the origin and acquisition of the streptomycin resistance determinant in Psa biovar 2, it is necessary to investigate resistance in the soil, rhizosphere, and phyllosphere microbiome. The comparative sequencing analyses of resistance genes may provide clues to understanding possible relationships such as horizontal gene transfer between Psa and coinhabiting streptomycin-resistant microorganisms.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by a Research promotion program of SCNU.

