Isolation of Nine Bacteriophages Shown Effective against Erwinia amylovora in Korea
Article information
Abstract
Erwinia amylovora is a devastating bacterial plant pathogen that infects Rosaceae including apple and pear and causes fire blight. Bacteriophages have been considered as a biological control agent for preventing bacterial infections of plants. In this study, nine bacteriophages (ΦFifi011, ΦFifi044, ΦFifi051, ΦFifi067, ΦFifi106, ΦFifi287, ΦFifi318, ΦFifi450, and ΦFifi451) were isolated from soil and water samples in seven orchards with fire blight in Korea. The genetic diversity of bacteriophage isolates was confirmed through restriction fragment length polymorphism pattern analysis. Host range of the nine phages was tested against 45 E. amylovora strains and 14 E. pyrifoliae strains and nine other bacterial strains. Among the nine phages, ΦFifi044 and ΦFifi451 infected and lysed E. amylovora only. And the remaining seven phages infected both E. amylovora and E. pyrifoliae. The results suggest that the isolated phages were different from each other and effective to control E. amylovora, providing a basis to develop biological agents and utilizing phage cocktails.
Erwinia amylovora is a plant pathogen that causes fire blight in the family Rosaceae. E. amylovora infection results in economic losses, and was first reported in the United States. Fire blight has spread in many countries in North America, New Zealand, and Europe (Gayder et al., 2019). In 2015, E. amylovora was first reported in a pear orchard of Anseong in Korea (Park et al., 2016). Since 2018, the occurrence of E. amylovora infections have increased rapidly in Korea. A total of 348 orchards were buried as a public control measure from 2015 to 2019 (Ham et al., 2020).
Antibiotics, copper compounds, and microbes are commonly used to control fire blight. In Korea, E. amylovora has been controlled by antibiotics such as kasugamycin, oxytetracycline, and streptomycin, copper compounds such as copper hydroxide, copper sulfate basic, oxine copper, and tribasic copper sulfate, and microbes such as Bacillus subtilis (Lee et al., 2018). However, use of antibiotics led to the emergence of resistant bacteria in several countries and its accumulation in the environment and in humans (Aćimović et al., 2015).
With the reporting of resistant bacteria and the expansion of the restriction on the use of antibiotics in agriculture, interest in biological controls considered environmentally friendly is increasing. Biological controls using bacteria, fungi, and bacteriophages have been developed. Antagonistic bacteria such as Pseudomonas, Pantoea, Enterobacter, Serratia, and Bacillus genera were reported for control of fire blight to suppress E. amylovora. For example, Pantoea agglomerans, B. subtilis, Bacillus amyloliquefaciens, and Pseudomonas fluorescens were registered bacteria products and commercially available (Dagher et al., 2020; Mechan Llontop et al., 2020; Mikiciński et al., 2016; Sharifazizi et al., 2017). Recently, use of phage therapy as biological control has been emphasized, in which bacteriophages are expected to prevent bacterial infections. Bacteriophages are viruses that replicate themselves and consist of nucleic acids and proteins. Additionally, bacteriophages are present in the surrounding environment of soil, air, and water and infect only target bacteria without any negative effects on humans or plants. Accordingly, phage therapy is non-toxic, eliminating just target bacteria, and requires small doses (Essa et al., 2020; Loc-Carrillo and Abedon, 2011; Principi et al., 2019).
Although bacteriophage use may lead to the emergence of resistant bacteria, and some of them have a narrow host range, combination of the phages should solve the problem. According to studies, combination of phages reduces the occurrence of phage-resistant bacteria as well as the bacterial population (Doss et al., 2017; Gayder et al., 2020). Born et al. (2011) reported that phage cocktails (L1/Y2 and L1/S6) are effective in controlling bacterial growth. Synergistic effects of phage cocktails is expected to affect different receptors of bacteria. When phages infect host bacteria, receptor binding proteins (RBPs) of phages bind to host receptors. Because of phage-host interactions, host specificity of phages varies at the level of genus, species, or even strains of host. Therefore, RBPs of phages are important to infect host bacteria (Dunne et al., 2018; Ge et al., 2020; Stone et al., 2019).
The key aspect of phage therapy to control fire blight is securing bacteriophages for all strains of target bacteria, E. amylovora, and use of combinations suppresses the appearance of resistant bacteria. In this study, the lytic activities, host specificity, and genetic diversity of nine bacteriophages isolated from soil and water samples in orchards that showed fire blight disease were evaluated.
To obtain E. amylovora-specific phages in Korea, environmental samples were collected from apple and pear orchards in which fire blight was observed in 2020. Each sample (10 ml or 10 g) was mixed with 10 ml of tryptic soy broth (TSB) and 1 ml of E. amylovora strain YKB 14808 (approximately 109 colony-forming unit [cfu]/ml). The samples mixed with E. amylovora were incubated overnight in a shaking incubator for enrichment of bacteriophages. All samples were centrifuged at 10,000 ×g for 10 min at 4°C and filtered using a 0.22 μm PVDF syringe filter (Millipore, Bedford, MA, USA) for removal of bacteria cells and other debris. Dotting assays and plaque assays were carried out as described previously with minor modifications (Park et al., 2018). Presence of bacteriophages was confirmed by dotting assays. When 4 ml of soft agar (TSB containing 0.4% agar) was heated by microwave and cooled to 40°C, 20 μl of overnight cultured E. amylovora strain YKB 14808 was mixed and poured onto tryptic soy agar (TSA) plates. After the soft agar layer was solid, 10 μl of the environmental samples incubated with E. amylovora were added and incubated at 28°C overnight. Based on dotting assay results, bacteriophages were purified by plaque picking and passaged by plaque assays at least five times. In plaque assays, 4 ml of soft agar, 20 μl of overnight cultured E. amylovora strain YKB 14808, and 100 μl of bacteriophage (103 plaque-forming unit [PFU]/ml) were mixed and poured on TSA. After incubation at 28°C overnight, differently sized and shaped single plaques were isolated. The 29 environmental samples were collected from nine orchards of three cities to isolate bacteriophages with genetic diversity. Among them, nine bacteriophages were isolated from seven samples. Information of samples and isolated bacteriophages is listed in Table 1. Three bacteriophages (ΦFifi011, ΦFifi106, and ΦFifi287) were isolated from soils and three bacteriophages (ΦFifi044, ΦFifi450, and ΦFifi451) were isolated from the water in the pear orchard in Anseong. One bacteriophage,ΦFifi051, was isolated from soils and two bacteriophages (ΦFifi067 and ΦFifi318) were isolated from water in the apple orchard in Chungju. Plaques of bacteriophages are shown in Fig. 1. Two phages (ΦFifi044 and ΦFifi451) formed big and clear plaques. The plaque diameter of two phages ranged from 2 to 4 mm. Seven phages (ΦFifi011, ΦFifi450, ΦFifi051, ΦFifi067, ΦFifi106, ΦFifi287, and ΦFifi318) formed smaller but clear plaques. The plaque diameter of seven phages irregularly ranged from 0.5 to 2 mm The plaque diameter of previously reported Erwinia phages generally range from 0.5 to 4 mm; phage phiEa2809 formed variable sized plaques (0.5–2 mm), H5K and phiEaP-8 formed plaques with an average diameter of 1–2 mm, and Pea1 formed plaques with an average diameter of 2–4 mm (Lagonenko et al., 2015; Nagy et al., 2015; Park et al., 2018; Ritchie and Klos, 1977).
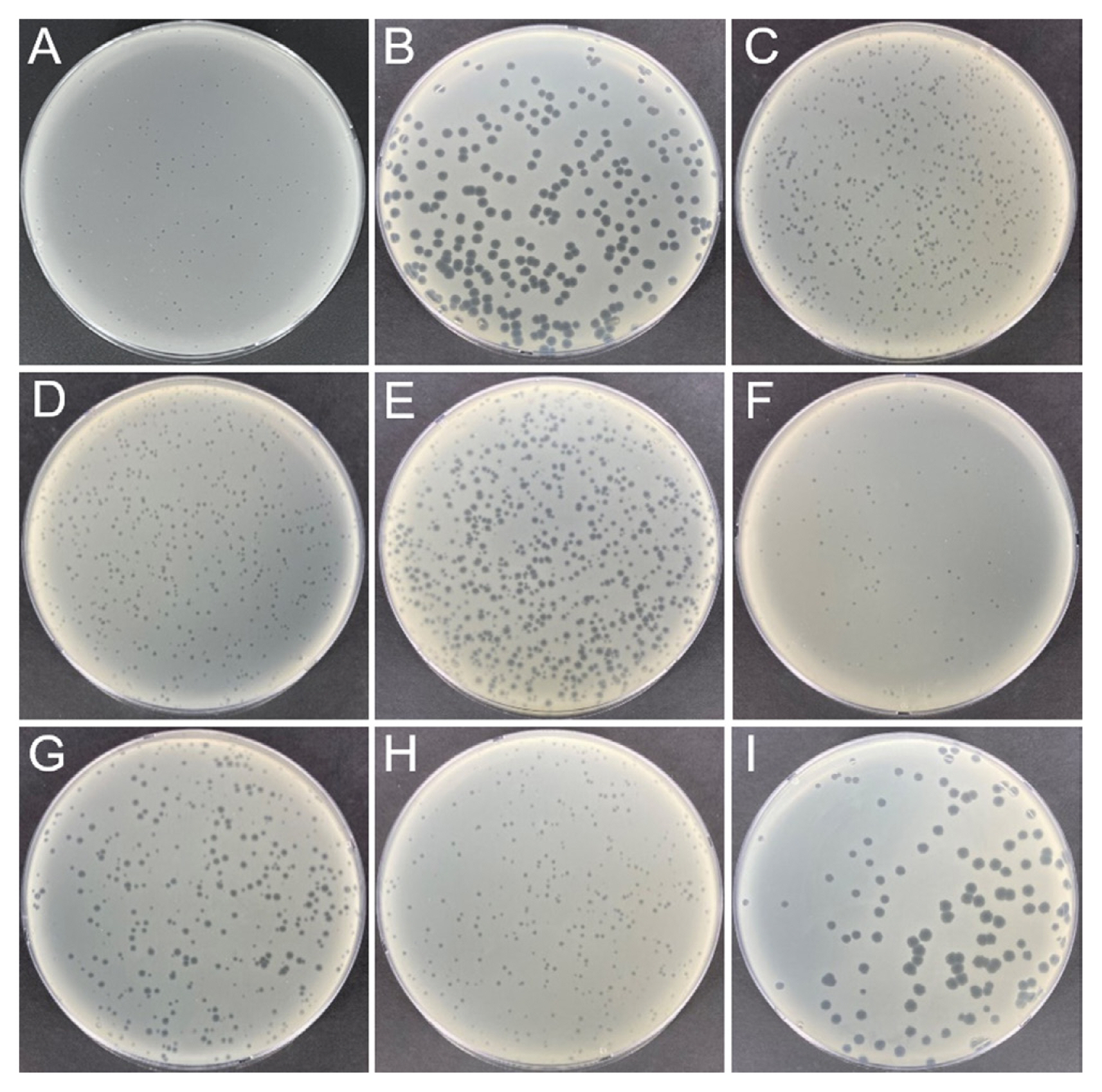
Plaque morphology of nine bacteriophages against Erwinia amylovora strain YKB 1480: (A) ΦFifi011, (B) ΦFifi044, (C) ΦFifi051, (D) ΦFifi067, (E) ΦFifi106, (F) ΦFifi287, (G) ΦFifi318, (H) ΦFifi450, (I) ΦFifi451.
To examine the genetic diversity of bacteriophages, genomic DNA of nine bacteriophages was extracted with a phage DNA isolation kit (Norgen Biotek, Thorold, ON, Canada) according to the manufacturer’s instructions. One μg genomic DNA was digested with restriction enzyme AseI (NEB, Ipswich, MA, USA) or HindIII (Takara, Tokyo, Japan) in a final volume of 20 μl at 37°C for 2 h. Two μl of LoadingSTAR (Dyne Bio Inc., Seongnam, Korea) was mixed with 10 μl of samples and loaded on an 0.7% agarose gel (70 V, 2 h). Restriction fragment length polymorphism (RFLP) analysis of the nine bacteriophages is shown in Fig. 2. All phages showed different DNA patterns in AseI except for ΦFifi044 and ΦFifi451. Because ΦFifi044 and ΦFifi451 showed similar RFLP patterns by AseI, the DNA patterns of two phages were additionally compared by HindIII. The two phages showed different DNA patterns. And clustering based on RFLP pattern for the nine bacteriophages were carried out. The difference of nine phages was confirmed through the dendrogram depicting clustering in RFLP of nine bacteriophages based on Neighbor joining generated in ToalLab Quant v.13.2 algorithm (Supplementary Fig. 1).
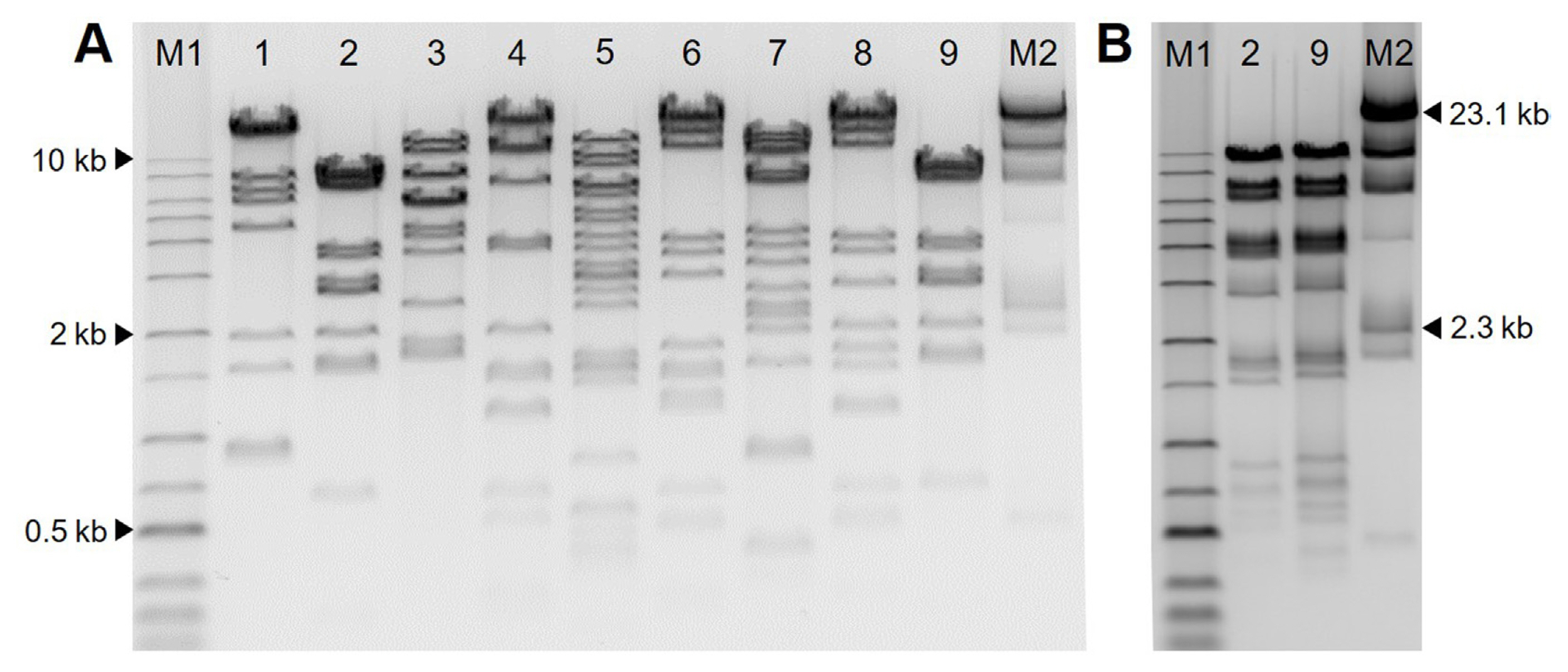
Restriction enzyme digestion pattern of nine bacteriophages. Genomic DNA from bacteriophages are extracted and digested with AseI for all phages and with HindIII for ΦFifi044 and ΦFifi451 for 2 h. (A) Agarose gel electrophoresis restriction patterns of DNA digested with AseI. Lane M1, 1 kb plus ladder (Dyne Bio Inc., Seongnam, Korea); lane 1, ΦFifi011; lane 2, ΦFifi044; lane 3, ΦFifi051; lane 4, ΦFifi067; lane 5, ΦFifi106; lane 6, ΦFifi287; lane 7, ΦFifi318; lane 8, ΦFifi450; lane 9, ΦFifi451; lane M2, Lambda/HindIII marker (Bioneer, Daejeon, Korea). (B) Agarose gel electrophoresis restriction patterns of DNA digested with HindIII. Lane M1, 1 kb plus ladder; lane 2, ΦFifi044; lane 9, ΦFifi451; lane M2, Lambda/HindIII marker.
To confirm the host range of the nine bacteriophages, E. amylovora strains, closely related E. pyrifoliae strains and other bacteria were used. E. amylovora and E. pyrifoliae were isolated from fire blight-infected apple and pear trees in diverse locations of Korea from 2015 to 2020 (Anseong, Asan, Chungju, Cheonan, Cheorwon, Chuncheon, Eumseong, Goseong, Gwangju, Hongcheon, Hwacheon, Icheon, Iksan, Jecheon, Jincheon, Mungyeong, Paju, Pocheon, Yanggu, and Yeoncheon) (Supplementary Table 1). All Erwinia strains were identified based on polymerase chain reaction (PCR) with species-specific primers, and identifications were confirmed using 16S rRNA sequence analysis. E. amylovora strains were identified by PCR by amplifying pEA29 using A (5′-CGGTTTTTAACGCTGGG-3′)/B (5′-GGGCAAATACTCGGATT-3′) primers (Bereswill et al., 1992). PCR conditions were as follows. Initial denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 50 s, and a final extension at 72°C for 7 min followed by 30 cycles for pEA29. E. pyrifoliae strains were identified by PCR by amplifying cps genes using CPS1 (5′-CGCGGAAGTGGTGAGAA-3′)/CPS2c (5′-GAACAGATGTGCCGAGTA-3′) primers (Kim et al., 2001). PCR conditions were as follows. Initial denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 50 s, and a final extension at 72°C for 7 min followed by 30 cycles for cps genes. PCR amplification was conducted using SafeDry Taq PCR Premix (CellSafe, Yongin, Korea) with primers in a final volume of 20 μl. For confirming the identification of bacteria, DNA from isolates was amplified by PCR using 16S rRNA universal primers 1492R (5′-GGTTACCTTGTTACGACTT-3′)/27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and sequenced using 16S rRNA universal primers 907R (5′-RCCGTCAATTCMTTTRAGTTT-3′)/785F (5′-GGATTAGATACCCTGGTA-3′) followed by Macrogen Co. (Seoul, Korea). The nine isolated bacteriophages were examined in regard to their host range through dotting assays against 45 strains of E. amylovora, 14 strains of E. pyrifoliae, and nine other bacteria (Dickeya zeae, Escherichia coli, P. agglomerans, Pectobacterium carotovorum, P. fluorescens, Xanthomonas campestris, Bacillus thuringiensis, Enterococcus faecalis, and Staphylococcus xylosus). Bacteria were grown in TSA or TSB at 28°C, but E. coli strain ATCC 43895, E. faecalis strain ATCC 35038, and S. xylosus strain ATCC 29971 were grown in TSA or TSB at 37°C. A volume of 4 ml of soft agar mixed with 20 μl of overnight cultured E. amylovora strain YKB 14808 was poured on agar plates and 10 μl bacteriophage aliquots (108 PFU/ml) were added and incubated overnight. The results of host range tests of the nine phages are shown in Fig. 3. ΦFifi044 and ΦFifi451 were only effective against E. amylovora strains. On the other hand, the other seven phages were effective against both E. amylovora and E. pyrifoliae strains. All nine phages were not effective against other tested bacterial strains. Erwinia phages reported so far can be divided into two groups according to the host range. The first is a group with a narrow host range specific to E. amylovora, such as Pea1, 2, 5, and Hena1 (Besarab et al., 2020; Ritchie and Klos, 1977). The second group includes phages with a wide host range, including the Pantoea species. Phage phiEa2809, vB_EamM_Y3, Deimos-Minion, Simmy50, RAY, Special G, Bosolaphorus, Desertfox, MadMel, and Mortimer are broad host range phages infecting E. amylovora, P. agglomerans, and Pantoea vagans strains (Buttimer et al., 2018; Lagonenko et al., 2015). Erwinia and Pantoea species belong to Erwiniaceae bacteria. Therefore, most reported phages were tested for host range against Erwina and Pantoea strains (Sharma et al., 2019). In Korea, four E. amylovora bacteriophages were reported. phiEaP-8, with a narrow host range, is effective against only E. amylovora and E. pyrifoliae strains (Park et al., 2018). Phage pEa_SNUABM_12, 47, and 50, with a broad host range, is effective against E. amylovora, E. pyrifoliae, Erwinia spp., and Serratia maarcescens strains (Kim et al., 2020). ΦFifi044 and ΦFifi451 infected and lysed E. amylovora only. This is the first report on the isolation of E. amylovora-specific bacteriophages in Korea. The other seven phages were specific to E. amylovora and E. pyrifoliae, similar to some previously reported phages. E. pyrifoliae was reported and classified as a novel species causing black shoot blight disease of pear and apple in Korea, and the symptoms are similar to those of fire blight (Kim et al., 1999). In Korea, these phages may be useful as biological control agents because E. amylovora and E. pyrifoliae can coexist in the same environment.

Heatmap showing the host range of nine isolated phages. Susceptibility of bacteriophages against Erwinia amylovora, E. pyrifoliae, and other bacterial strains is indicated by a color scale, from white to red (clearing of 1 to 4; 0 = no lysis).
Control of E. amylovora infections is necessary to protect apples and pears and reduce economic losses. Chemical controls such as antibiotics and copper compounds have the potential to affect the environment and humans by giving rise to resistant bacteria as well as inhibiting the growth of all microorganisms to which they are exposed, including those beneficial to plants. Bacteriophages are considered a suitable biological control because of their host specificity and environmental friendliness. Therefore, nine bacteriophages effective against E. amylovora were isolated and characterized regarding plaque morphology, genetic diversity and host range. The results of gDNA pattern analysis show that the nine phages are different from each other, and this diversity is considered a positive factor for the synergistic effect of a phage cocktail. According to the host range and plaque morphology, the nine phages are divided into two groups. In particular, ΦFifi044 and ΦFifi451 belonging to the first group are noteworthy in that they are the first E. amylovora specific phages reported in Korea.Further studies will provide information on the characteristics of phages for application as biocontrol agents.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
We thank Dr. Young-Kee Lee for providing the YKB strains of E. amylovora and E. pyrifoliae. This work was supported by the Rural Development Administration (RDA; grant number PJ01496501).
Notes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.

