Changes in the Composition and Microbial Community of the Pepper Rhizosphere in Field with Bacterial Wilt Disease
Article information
Abstract
Bacterial wilt caused by Ralstonia solanacearum is considered one of the most harmful diseases of pepper plants. Recently, research on plant disease control through the rhizosphere microbiome has been actively conducted. In this study, the relationship with disease occurrence between the neighboring plant confirmed by analyzing the physicochemical properties of the rhizosphere soil and changes in the microbial community. The results confirmed that the microbial community changes significantly depending on the organic matters, P2O5, and clay in the soil. Despite significant differences in microbial communities according to soil composition, Actinobacteriota at the phylum level was higher in healthy plant rhizosphere (mean of relative abundance, D: 8.05 ± 1.13; H: 10.06 ± 1.59). These results suggest that Actinobacteriota may be associated with bacterial wilt disease. In this study, we present basic information for constructing of healthy soil in the future by presenting the major microbial groups that can suppress bacterial wilt.
Ralstonia solanacearum is a species complex and is a phytopathogenic bacterium that causes severe bacterial wilt in more than 50 families and 450 plant species (Denny, 2006). Bacterial wilt caused by R. solanacearum causes significant loss of economically important crops in tropical, subtropical, and temperate regions (Hayward, 1991). It is also known that pathogens survive for long periods in water or soil and can cause disease outbreaks when environmental conditions are favorable for infection (Coutinho, 2005; Grey and Steck, 2001; van Elsas et al., 2001). Therefore, long-term survival of R. solanacearum can be a threat to crop cultivation, making it difficult to manage bacterial wilting. Despite the crop production risks of R. solanacearum, there are no effective management practices for bacterial wilting, and chemical control cannot be used to completely eliminate bacterial pathogens from soil and water. The cultivation of resistant varieties can be presented as a good alternative, but it takes a long time to develop resistant varieties (Saddler, 2005).
Recently, microbes surrounding plants have been shown play an important role in controlling plant diseases (Berendsen et al., 2018; Kwak et al., 2018; Lee et al., 2021; Mendes et al., 2011). In the case of persistent and serious plant diseases, soil after disease outbreaks also exhibits the ability to inhibit the progression of plant diseases (Weller et al., 2002). Soils with these characteristics are called “disease-suppressive soils” and are defined as soils that can suppress the onset of disease despite the coexistence of pathogens and susceptible plant hosts (Cook and Baker, 1983). These disease-suppressive soils are represented by a variety of pathogens, including fungi and bacteria such as R. solanacearum (Klein et al., 2013; Kobayashi and Komada, 1995; Mendes et al., 2011; Rosenzweig et al., 2012; Shiomi et al., 1999).
The soil microbial community plays an important role in disease-suppressive soil function (Mendes et al., 2011). Recent advances in techniques related to microbiome analysis have confirmed that disease-suppressing soils are achieved through complex microbial community balance. In particular, Pseudomonas, Bacillus, and Streptomyces have been found to be important bacterial genus group in the microbial community of the plant rhizosphere (Abbasi et al., 2021; Cha et al., 2016; Kloepper et al., 2004; Lee et al., 2021; Raaijmakers and Weller, 1998).
It has also been recently reported that the disruption of the balance of the microbial community in plants is associated with the development of plant diseases (Lee et al., 2021). In the case of Arabidopsis phyllosphere, it was confirmed that the dysbiosis of the microbial community due to the decrease in the relative abundance of Firmicute affects the health of plants (Chen et al., 2020). In the plant rhizosphere, tomato plants that developed blight showed a decrease in Firmicutes and Actinobacteria. In addition, the destruction of artificial Firmicutes and Actinobacteria in the microbial community using vancomycin was confirmed to be susceptible to the disease (Lee et al., 2021).
In this study, we compared plant rhizosphere microbial communities with and without bacterial wilt disease in pepper fields at two different geographic locations in Korea. Analysis of rhizosphere samples using a 16S rRNA amplicon sequencing-based approach revealed that the neighboring plant rhizosphere microbiome differs according to the presence (D) or absence (H) of disease and pathogens. These results suggest the possibility that changes in the Actinobacteria and phosphate content in the pepper rhizosphere soil are key factors in inducing or inhibiting bacterial wilt. Our results provide basic information on the pathogenesis of plants and will help understand the approach of microbial communities as biological control agents based on biodiversity and balance.
Pepper rhizosphere samples with and without bacterial wilt were collected from three farms in Imsil (4, 5, 6: 35°42′20.63″N, 127°17′13.77″S) and four farms (10, 11: 35°22′31.72″N, 127°12′26.64″S; 13, 15: 35°22′47.70″N, 127°06’12.94″S) in Sunchang, Korea. Pepper plants that developed bacterial wilt showed signs of withering in the entire plant. In the case of a tomato sample that did not develop bacterial wilt, the prepared plant was collected immediately next to the diseased plant, and there were no symptoms.
Bacterial wilt symptoms appeared randomly in the field, and it was confirmed that healthy plants were adjacent to the plants with bacterial wilt (Fig. 1). When R. solanacearum was tested in the rhizosphere of peppers, pathogens were identified in diseased peppers, whereas pathogens were not cultured in healthy peppers (data not shown). Therefore, it was confirmed that the pathogenic bacteria did not spread from the rhizosphere of a diseased plant to the rhizosphere of a healthy plant during the disease outbreak period.

Sampling of adjacent pepper plant rhizosphere soils. A map of Seven pepper farms (4, 5, 6, 10, 11, 13, and 15) in two regions (Imsil and Sunchang) where sampling was performed. Images of neighboring healthy plants (H) and diseased plants (D).
For rhizosphere soil samples, large soil particles were removed from the roots and only rhizosphere soils bound to plant roots were collected. The collected plant roots were suspended in sterile distilled water for 30 min, and the soil suspension was centrifuged at 8,000 rpm for 10 min. Soil pellets containing the microbiome were stored at −80°C until the next experiment.
To analyze the physical and chemical properties of the collected pepper rhizosphere soil, nitrogen (%), organic matter (g/kg), P2O5 (mg/kg), potassium (cmol+/kg), calcium (cmol+/kg), sodium in the soil (cmol+/kg), sand (%), silt (%), and clay (%) were analyzed by the Korea Agriculture Technology Promotion Agency (KOAT, Iksan, Korea). The physicochemical properties of the rhizosphere soil were compared with each sample using principal component analysis (PCA). PCA of rhizosphere soils collected from two regions and seven fields confirmed the differences between samples. Consequently, it was confirmed that they were divided into three groups based on the collected regions. Specifically, it was confirmed that soil collected from Imsil (D6 and H6) had characteristics similar to those of the Sunchang sample. However, the soil composition was not divided according to the presence or absence of disease (Fig. 2A).
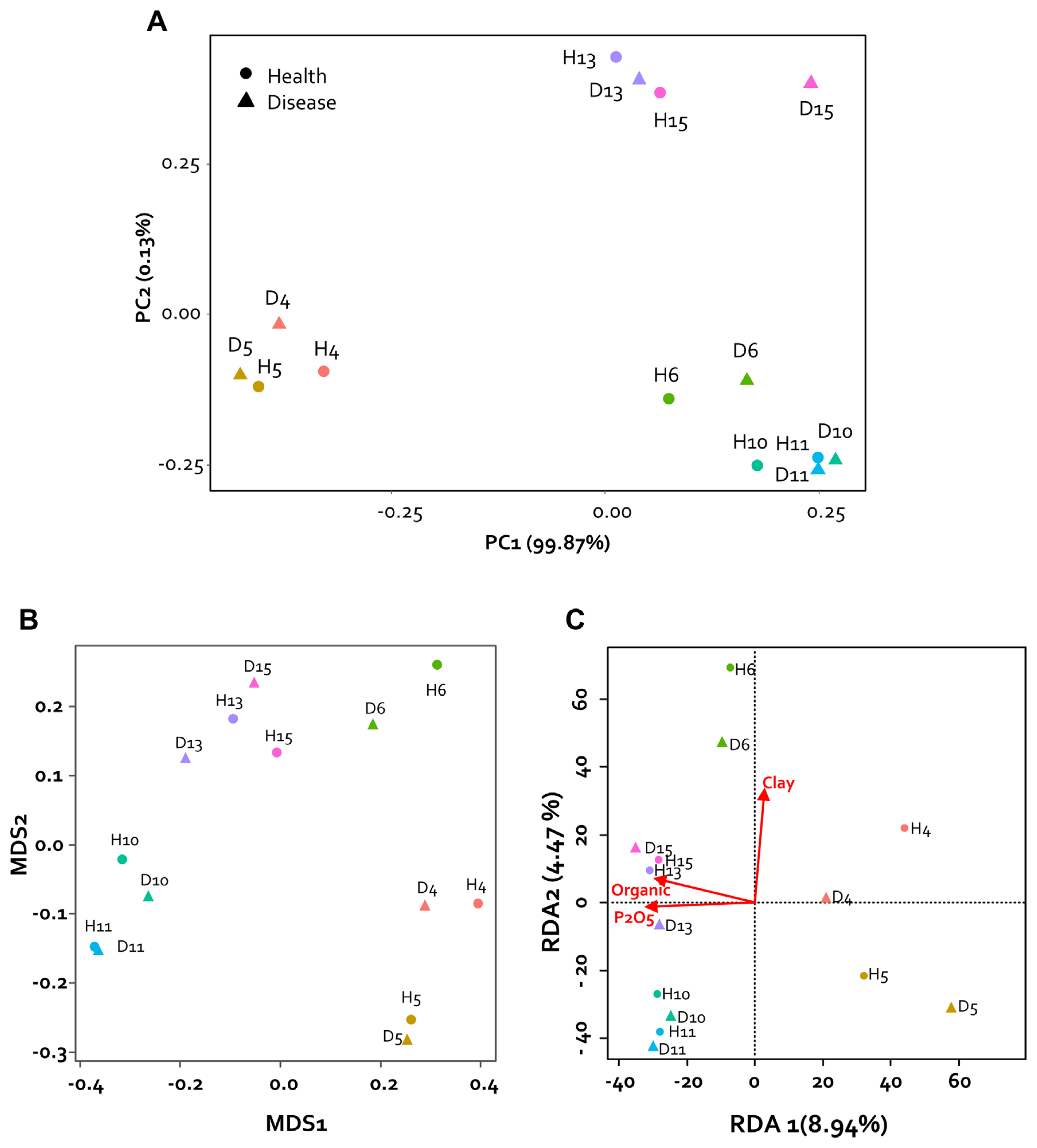
Physicochemical principal component analysis (PCA) and microbiome analysis of rhizosphere soil. (A) PCA was performed based on nitrogen (%), organic matter (g/kg), P2O5 (mg/kg), potassium (cmol+/kg), calcium (cmol+/kg), sodium in the soil (cmol+/kg), sand (%), silt (%), and clay (%). (B) Non-metric multidimensional scaling (NMDS) alignment of the variation in the bacterial community structure of the pepper rhizosphere according to region and disease occurrence. Data represent arrays based on Bray-Curtis distances between 14 samples. Samples (points) are colored according to region. The shape indicates the presence or absence of disease (triangle for disease; circle for health). (C) Pepper rhizosphere microbiome sensitivity to environmental parameters. Redundancy analysis (RDA) was used to evaluate the impact of environmental fluctuations on microbial community composition with and without disease outbreaks across regions. The total variance (percentage) explained by each axis is shown in parentheses. Environmental factors (Organic, P2O5, and Clay) significantly explained the observed compositional changes in the microbial community.
Therefore, in order to confirm the difference in the microbial community in the rhizosphere soil, microbial genomic DNA was extracted from the harvested rhizosphere soil using the DNeasyPowerSoil Kit (Qiagen, Germantown, MD, USA), and DNA was quantified using amicroplate reader (Infinite M200PRO, TECAN, Männedorf, Switzerland), PicoGreen (Turner Biosystems, Sunnyvale, CA, USA), and Nanodrop (Thermo Fisher Scientific, Wilmington, DE, USA). The V3 and V4 regions of the 16S rRNA gene were amplified using polymerase chain reaction (PCR) and used for microbiome analysis (Lundberg et al., 2013). The PCR products were separated by electrophoresis on a 1% agarose gel and visualized using a Gel-Doc system (Bio-Rad, Hercules, CA, USA).
The PCR products were then purified using the CleanPCR Kit (CleanNA, Waddinxveen, Netherlands) and the same concentrations of the purified products were pooled together. The CleanPCR Kit was used to remove off-target short fragments and the quality and size of the PCR products were evaluated using a DNA 7500 chip from a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA). The pooled amplicons were sequenced at Macrogen (Seoul, Korea) using the Illumina MiSeq platform according to the manufacturer’s instructions.
The basic sequence quality of the amplified genes was assessed using FastQC and low-quality cutoffs for forward and reverse reads were determined. The reads were then imported forward and backward into QIIME2 (v2022.2) (Bolyen et al., 2019) for quality control, and diversity analysis. Sequencing was performed with MiSeq platform. The sequence noise and chimera removal by using DADA2 (Callahan et al., 2016). The operational taxonomic unit (OTU) table was normalized by dividing the reads per OTU for each sample by the sum of the reads for that sample.
Non-metric multidimensional scaling analysis was performed to detect the difference in microbial composition between healthy pepper rhizosphere and diseased rhizosphere samples collected from seven farms in two regions of Korea. As a result, no group was clustered based on the presence or absence of disease within the region (Fig. 2B). However, it was confirmed that the composition of the microbial community of each sample shown in the main coordinate analysis was similar to the component analysis result of the collected sample. Therefore, through redundancy analysis analysis, the factors of soil composition were confirmed by the differences among the regions in the sample. It was confirmed that organic matter, P2O5, and clay played an important role in the formation of the microbial community (Fig. 2C). Therefore, it could be confirmed that the change in the microbiome by fertilizer and compost treatments was greatly affected. Nevertheless, the high relative abundance of Actinobacteria identified in our study in healthy plants is consistent with the previously reported results of increased wilting due to decrease in Firmicutes and Actinobacteria (Lee et al., 2021). On the other hand, our results have the characteristic that pathogenic bacteria are not observed in the disease pepper rhizosphere. These results seems that our health samples reflect the microbial community before the invasion of pathogens.
Furthermore, to confirm the difference in microbial composition between healthy and diseased rhizospheres, four regional microbial communities were averaged by calculate the mean of the relative abundance, and relative abundance analysis was performed accordingly. The classification levels of all sample reads were assigned to the species level using the Silva 138 Reference Taxonomy Database (https://docs.qiime2.org/2022.2/data-resources/). Relative abundance analysis was performed at the class level, and Gammaproteobacteria (D: 19%, H: 18%), Alphaproteobacteria (D: 14%, H: 15%), Bacteroidia (D: 7.8%, H: 8.7%), and Actinobacteria (D: 5.5%, H: 7.2%) were the major bacterial communities (Fig. 3A). When comparing the read numbers for each of the five main sentences, there were no statistically significant differences.
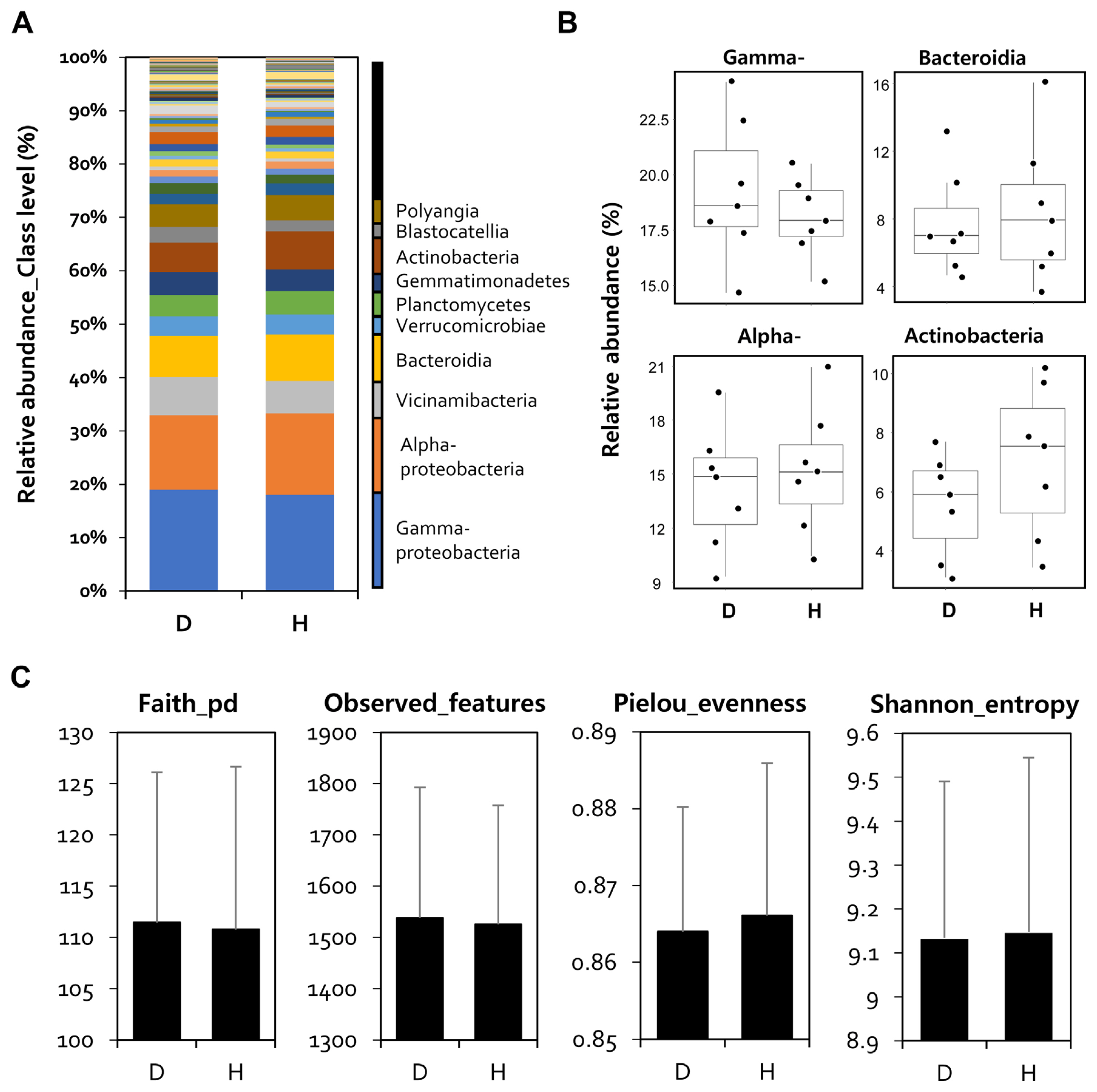
Bacterial community structure in pepper rhizosphere. (A) A percentage of bacterial community abundance at class level in both soil groups with (D) and without (H) disease outbreaks of pepper rhizosphere. Relative abundance is calculated by averaging the abundance of duplicate samples from each soil group in the pepper rhizosphere. (B) Differences in abundance of major microbiome in pepper rhizosphere microbiome. Data is displayed as a relative abundance (%). Each point corresponds to a single sample. (C) Changes in the alpha diversity of the pepper rhizosphere microbiome.
However, in the case of Gammaproteobacteria, it increased in the rhizosphere of infected peppers, and Actinobacteria increased in healthy plants (Fig. 3B). Alpha diversity analysis with faith_Pd, features, Pielou_evenness, and Shannon_entropy showed that there was no difference in bacterial uniformity and abundance indices between the diseased and healthy pepper rhizospheres (Fig. 3C).
Furthermore, because the class-level ratio change is related to the disease-generating condition of the plant, each OTU showing a different read number was compared. Random forest (Breiman, 2001), a machine learning method, was applied to the OTU level dataset to analyze the discriminant rhizosphere microbiome components of diseased and healthy pepper rhizosphere (Fig. 4). Differential OTU according to disease incidence were confirmed as distinct changes in the relative abundance (Fig. 4A). To analyze the data, we performed a one-way ANOVA and a two-tailed Student’s t test in the R program (Pinheiro et al., 2011). Differences between samples were considered statistically significant at P < 0.05. In particular, the two differential OTUs belonging to Myxococcia and Actinobacteriota showed 2.2-fold and 11-fold higher read numbers in healthy plant rhizosphere, respectively (Fig. 4B).
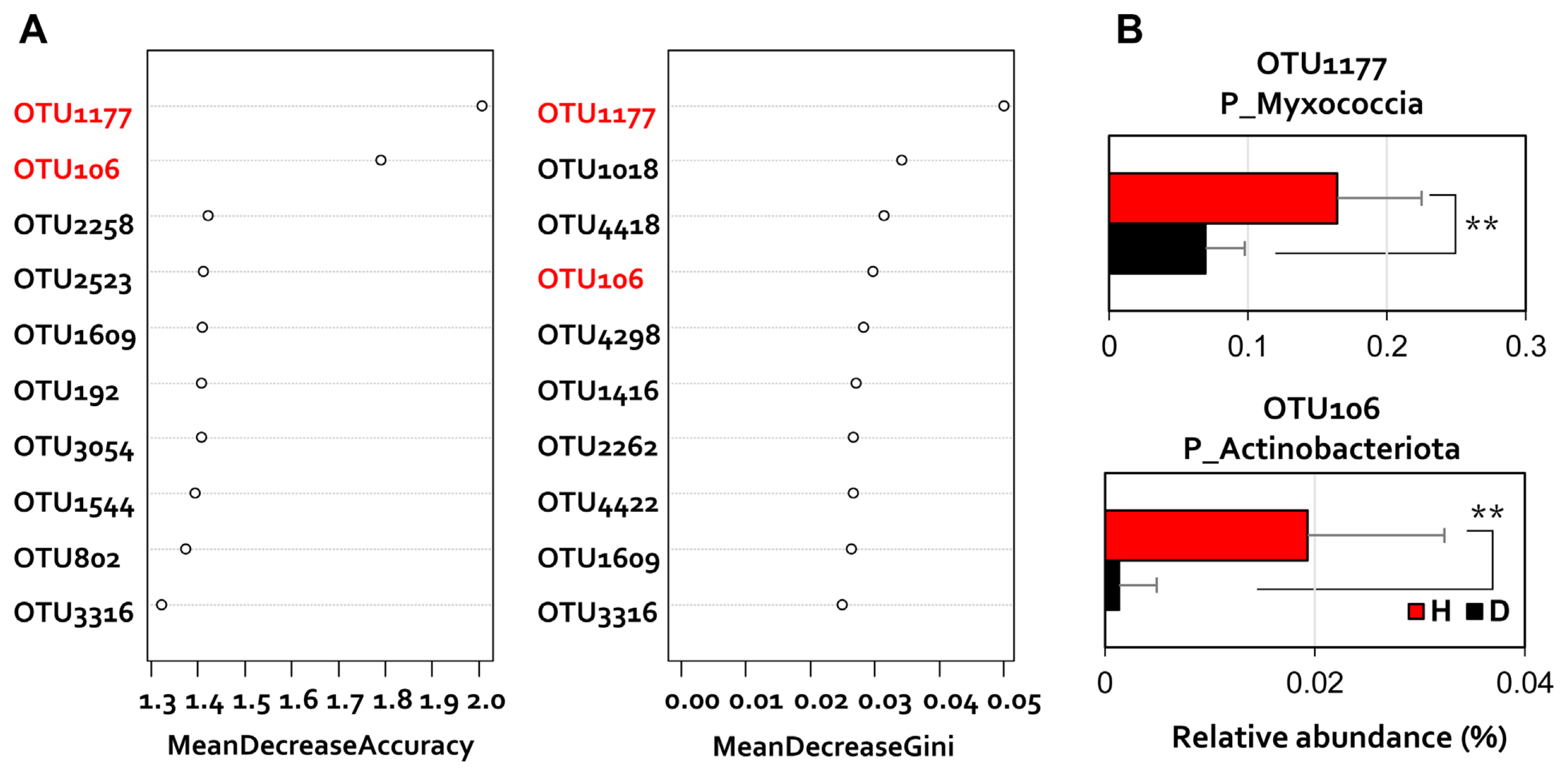
Random forest analysis. (A) The top 10 operational taxonomic units (OTUs) with the highest discriminant power among pepper rhizosphere groups by disease incidence are listed. (B) The red fonts show the highly relevant OTUs between the two groups. The relative abundance (%) at the phylum level of OTU1177 and OTU106 are highly related (healthy plants are shown in red; diseased plants are shown in black). **P < 0.01.
As a result of our study, it was confirmed that the microbial community in the rhizosphere changes according to the presence or absence of disease. It has been reported that plants manipulate the microbiome of the rhizosphere to suppress disease (Berendsen et al., 2018; Kwak et al., 2018; Lee et al., 2021; Mendes et al., 2011). In this study, Actinobacteria, which are relatively abundant in the healthy plant rhizosphere, are known to inhibit the growth of several plant pathogens in the plant rhizosphere, and produce extracellular enzymes that contribute to crop production and boosting immunity by decomposing complex mixtures in the soil (Bhatti et al., 2017; Kim and Kwak., 2022; Roy et al., 2019).
Because the homeostatic balance of microbial community composition is important for healthy host-microbial relationships, both the abundance and destruction of the microbiome serve as important mechanisms of disease pathogenesis in plants. Recent studies have mainly focused on the homeostatic balance of microbial communities by consortia of single and unidentified bacteria that are protected in the rhizosphere (Berendsen et al., 2018; Lee et al., 2021).
Consequently, it was confirmed the change of the Actinomycete community and organic matter, P2O5, and clay in the rhizosphere of pepper plants with bacterial wilt. Furthermore, it is necessary to understand the individual microbial determinants and study disease suppression through artificial consortium construction. Our results may provide a basis for the selection and development of plant health- and growth-promoting microbial consortia or biological control agents through the study of the microbiome following soil bacterial outbreaks.
Acknowledgments
This work was supported by a “Research Program for Agricultural Science & Technology Development (Project No. PJ01505101)” provided by the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
