Seed Transmission of Tomato yellow leaf curl virus in White Soybean (Glycine max)
Article information
Abstract
Tomato yellow leaf curl virus (TYLCV) infection of the common bean (Phaseolus vulgaris) has been reported, but soybean (Glycine max) has not previously been identified as a TYLCV host. Five cultivars of white soybean were agro-inoculated using an infectious TYLCV clone. At 30 days post-inoculation, they showed infection rates of 25% to 100%. Typical TYLCV symptoms were not observed in any inoculated plants. To examine whether TYLCV was transmitted in soybean seeds, DNA was isolated from bundles of five randomly selected seeds from TYLCV-inoculated soybean plants and amplified with a TYLCV-specific primer set. With the exception of one bundle, all bundles of seeds were verified to be TYLCV-infected. Virus dissemination was also confirmed in three of the 14 bunches. Viral replication was also identified in seeds and seedlings. This is the first report demonstrating that soybean is a TYLCV host, and that TYLCV is a seed-transmissible virus in white soybean.
Tomato yellow leaf curl virus (TYLCV) is a begomovirus that causes severe economic loss due to decreased crop production in tropical and subtropical regions (Czosnek and Laterrot, 1997; Moriones and Navas-Castillo, 2000). In Korea, TYLCV has circulated consistently across the country since its outbreak in 2008 (Kil et al., 2014a; Lee et al., 2010). TYLCV has a circular single-stranded DNA (ssDNA) genome about 2.8 kb in length, which is encapsidated in a twinned icosahedral virion (Moriones and Navas-Castillo, 2000). TYLCV-infected tomato plants exhibit severe symptoms such as curling and yellowing of young leaves and stunting, causing critical production loss in tomato cultivation (Papayiannis et al., 2011). In addition to tomato, other cultivated crops such as hot and sweet peppers (Capsicum annuum), cucurbit (Cucumis species), eustoma (Eustoma grandiflora), and the common bean (Phaseolus vulgaris) have been identified as TYLCV hosts, as well as some weeds (Anfoka et al., 2009; Cohen et al., 1995; Kil et al., 2014a, 2014b, 2015; Moriones and Navas-Castillo, 2000; Navas-Castillo et al., 1999; Reina et al., 1999).
Until recently, TYLCV was believed to be inoculated only by whitefly-mediated transmission or artificial inoculation with an infectious clone (Stanley et al., 2001). However, it was recently confirmed that TYLCV-infected tomato seeds could transmit infectious virus (Kil et al., 2016). In addition, seed transmission of other begomoviruses such as Sweet potato leaf curl virus (SPLCV) and Mung bean yellow mosaic virus (MYMV) was also reported in sweet potato and black gram (Kim et al., 2015; Kothandaraman et al., 2016). Seed transmission of TYLCV has also been verified in sweet pepper (unpublished data).
Soybean (Glycine max) is one of the major crops cultivated around the world. This plant is most commonly used as a source of edible oil and a protein source for chickens and pigs (Graham and Vance, 2003; Lee et al., 2011; Ross-Ibarra et al., 2007). In Korea, it has been widely cultivated in an area of about 75,000 ha (as of 2014), and Jeju, Muan, Goheung, and Paju are its main production areas. It has been used as a main ingredient in tofu and soy milk and used for the production of fermented foods such as soy sauce and soybean paste. A previous study in Korea found that soybean plants can be infected by Soybean mosaic virus (SMV), Soybean dwarf virus, Alfalfa mosaic virus (AMV), Cucumber mosaic virus (CMV), Cowpea mosaic virus (CPMV), Soybean yellow mottle mosaic virus (SYMMV), Soybean yellow common mosaic virus (SYCMV), and Peanut stunt virus (PSV) (Baek et al., 2012). Although TYLCV infection of the common bean (P. vulgaris) has been reported (Navas-Castillo et al., 1999), soybean (G. max) has not previously been identified as a TYLCV host. In a previous report of weed hosts for TYLCV, two soybean plants that arose in a tomato cultivating area were identified as TYLCV-infected (Kil et al., 2014b). However, this result was not confirmed in any subsequent study.
One aim of this study was to confirm soybean (G. max) is a TYLCV host, as mentioned in a previous study (Kil et al., 2014b). Five cultivars of white soybean (‘Jinpung,’ ‘Taegwang,’ ‘Seonpung,’ ‘Neulchan,’ and ‘Daepung’) were agro-inoculated using an infectious TYLCV clone prepared in a previous study (Kil et al., 2014a) and cultivated in an isolated plant cultivation chamber in Wanju in 2015 (Fig. 1A). One month after inoculation, DNA was extracted using a Viral Gene-spin™ Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) from leaf samples of 20 inoculated soybean plants, as described previously (Kil et al., 2014a). The extracted DNA was analyzed by PCR using 1 × AccuPower ® PCR Master Mix (Bioneer, Daejeon, Korea) with TYLCV-specific primer sets. Based on the PCR results, the infection rates of ‘Jinpung’ and ‘Taegwang’ were 85.7% (6/7) and 100% (4/4), respectively; the other soybean cultivars showed infection rates of 25.0% to 66.7% (Table 1, Fig. 1B). Typical TYLCV symptoms were not observed in any inoculated plants. Viral replication in TYLCV-infected soybean plants was assessed by strand-specific amplification of double-stranded viral DNA (dsDNA) using virion sense- and complementary sense-specific primer sets introduced in a previous study (Rodríguez-Negrete et al., 2014). First, strand-specific templates were produced with T4 DNA polymerase (Takara, Shiga, Japan) and viral-specific primers containing a 5′ TAG (AGTTTAAGAACCCTTCCCGC) (OCS-TAG [5′-AGTTTAAGAACCCTTCCCGCGGACTTTACATGGGCCTTCAC-3′] or OVS-TAG [5′-AGTTTAAGAACCCTTCCCGCGAAGGCTGAACTTCGACAGC-3′]) from single-stranded viral templates. Then, 2 μl of the first-strand reaction was mixed with 10 μl of 2 × AccuPower® PCR Master Mix (Bioneer), 1 μl of 10 μM specific primers (TAG, OVS, or OCS), and 6 μl of nuclease-free water. Thermocycling conditions included one cycle at 95°C for 30 s, 40 cycles at 95°C for 10 s, and 60°C for 15 s; reactions were performed in a T100™ thermal cycler (Bio-Rad, Hercules, CA, USA) (Fig. 2A). When geminiviral DNA is replicated normally by a rolling-circle replication mechanism, ssDNA is converted into dsDNA intermediates. These intermediates can be amplified using primers designed to specifically identify the presence or absence of each ssDNA and dsDNA strand (Gutierrez, 1999; Rodríguez-Negrete et al., 2014) (Fig. 2A). Strand-specific amplification of DNA isolated from PCR-confirmed TYLCV-infected soybean plants revealed that both virion sense and complementary sense ssDNA strands were identified in ‘Jinpung’ and ‘Taegwang’ (Fig. 2B). These results indicate that white soybean (G. max) is susceptible to TYLCV infection.

(A) Overall study scheme. (B) PCR analysis of DNA extracted from the leaves of Tomato yellow leaf curl virus (TYLCV)-inoculated white soybean plants. DNA was amplified with a TYLCV-specific primer set. Lane N, no template control; lane P, positive control of TYLCV-infected tomato leaves; lane 1, ‘Jinpung’; lane 2, ‘Taegwang’; lane 3, ‘Seonpung’; lane 4, ‘Neulchan’; and lane 5, ‘Daepung.’
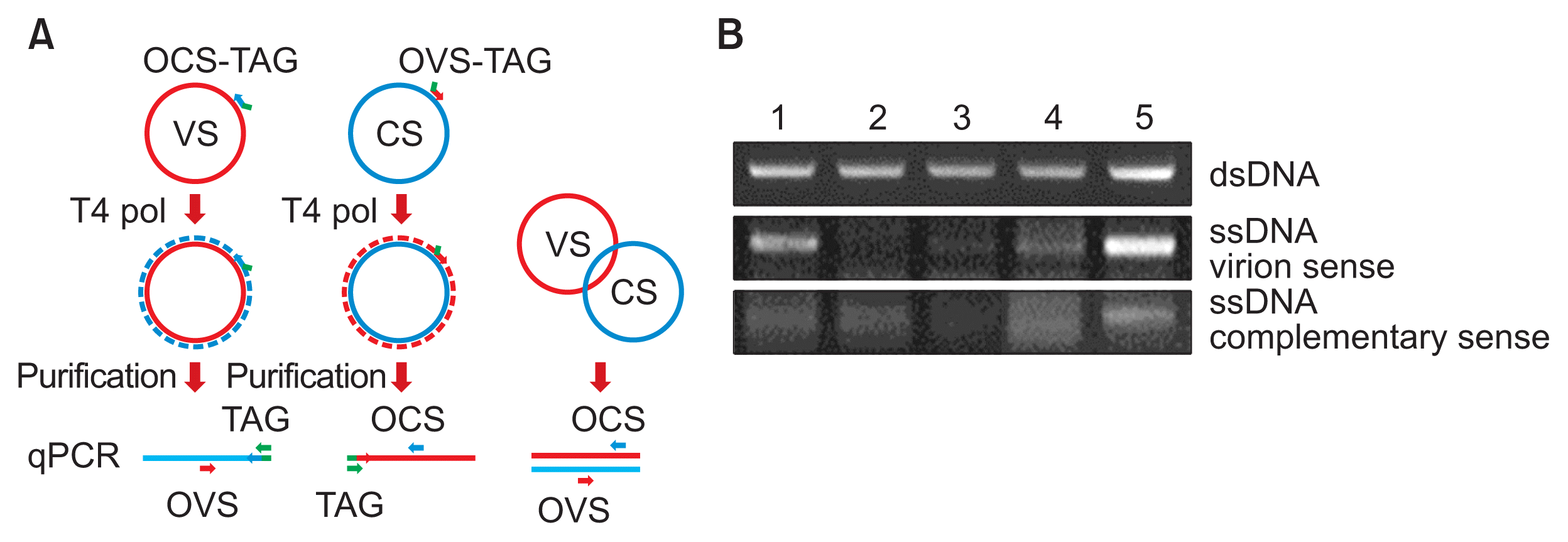
(A) Schematic representation of the two-step quantitative PCR (qPCR) procedure for the quantification of virion sense (VS) and complementary sense (CS) DNA molecules (modified from Rodríguez-Negrete et al., 2014). (B) Strand-specific amplification of DNA from seeds and seedlings using virion sense- and complementary sense-specific primer sets. Lane 1, ‘Jinpung’; lane 2, ‘Taegwang’; lane 3, ‘Seonpung’; lane 4, ‘Neulchan’; and lane 5, ‘Daepung.’ dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
To determine if TYLCV can be transmitted via the infected seeds of soybean, soybean seeds were harvested from TYLCV-infected plants. Collected seeds were surface-sterilized by exposure to 70% ethanol and 10% Clorox, followed by rinsing three times in sterile distilled water to prevent contamination (Trolinder and Goodin, 1988). Seeds were placed on a wet tissue (Wypall® L25 Wipers; Kimberly-Clark, Seoul, Korea) in a petri dish (SPL Life Sciences, Pocheon, Korea) and germinated in a plant bio low chamber (Dasol Scientific, Hwaseong, Korea) at 28°C (Fig. 3A). Five-day-old seedlings were collected for dissemination tests (Fig. 3A). DNA was isolated from bundles of five randomly selected soybean seeds and seedlings and then amplified with a TYLCV-specific primer set as described above. With the exception of one bundle of ‘Jinpung’ seeds, all bundles of seeds from infected soybean plants were verified as TYLCV-infected (Table 2, Fig. 3B). Virus dissemination was also confirmed in three of 14 bundles each containing five seedlings germinated from infected seeds (Table 2, Fig. 3B). Moreover, viral replication in seedlings of ‘Jinpung’ seeds was identified via strand-specific amplification (Rodríguez-Negrete et al., 2014) (Fig. 3C). These results confirm that TYLCV can be vertically transmitted via soybean seeds.
(A) Seeds and germinated seedlings from Tomato yellow leaf curl virus (TYLCV)-inoculated white soybean plants. (B) TYLCV-specific PCR analysis of seeds and seedlings from TYLCV-inoculated white soybean plants. Lane N, no template control; lane P, positive control of TYLCV-infected tomato leaves; lane 1, ‘Jinpung’; lane 2, ‘Taegwang’; lane 3, ‘Seonpung’; lane 4, ‘Neulchan’; and lane 5, ‘Daepung.’ (C) Strand-specific amplification of DNA from seedlings of ‘Jinpung’ soybeans germinated from TYLCV-infected seeds using virion sense- and complementary sense-specific primer sets. Lane D, double-stranded DNA; lane V, single-stranded DNA (virion sense); and lane C, single-stranded DNA (complementary sense).
Soybeans are used for a variety of applications and are cultivated worldwide. Moreover, many viruses have been reported to infect soybean plants (Graham and Vance, 2003; Lee et al., 2011; Ross-Ibarra et al., 2007). Of these viruses, Cowpea mild mottle virus (CPMMV), SMV, Tobacco ring spot virus (TRSV), and Tomato black ring virus (TBRV) have been reported to be transmitted via seeds (Athow and Bancroft, 1959; Brunt and Kenten, 1973; Kendrick and Gardner, 1924; Lister, 1960). Recently, Soybean vein necrosis virus (SVNV) was also identified as a seed-borne virus, making it the first Tospovirus to be implicated in seed transmission (Groves et al., 2016). Seed-borne viruses that can infect soybeans have been studied for a comparatively long time, because of their relatively high infection rates and the morphological characteristics of soybean seeds.
Based on results from a previous field survey (Kil et al., 2014b), we confirmed that soybean is a TYLCV host by agro-inoculation of five white soybean plants. In addition, we verified that TYLCV-infected soybeans can transmit the virus to their seedlings. This is the first report to demonstrate that soybean (G. max) is a TYLCV host, and that TYLCV is a seed-transmissible virus in white soybean. These results confirm that TYLCV seed transmission is not limited to tomatoes; we note that TYLCV can also be transmitted in sweet peppers (unpublished data). Although no reports in Korea have yet identified economic or other harm caused by TYLCV infection to soybean, TYLCV-infected soybeans could act as a reservoir (alternative host), thereby decreasing production of other crops such as tomatoes. Also, as soybeans are one of the major crops cultivated globally, TYLCV seed transmission in soybeans is potentially relevant worldwide. Therefore, it is necessary to determine the potential of TYLCV infection and transmission via seeds for various varieties of soybeans, predict the economic damage, and develop a quarantine policy based on these predictions.
Acknowledgments
This research was supported by a grant (311058-05-5-HD140) from the Agricultural Biotechnology Development Program of the Ministry of Agriculture, Food, and Rural Affairs of the Republic of Korea, and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, & Future Planning (2016R1C1B2014391).