Characterization of a Strain of Malva Vein Clearing Virus in Alcea rosea via Deep Sequencing
Article information
Abstract
Malva vein clearing virus (MVCV) is a member of the Potyvirus species, and has a negative impact on the aesthetic development of Alcea rosea. It was first reported in Germany in 1957, but its complete genome sequence data are still scarce. In the present work, A. rosea leaves with vein-clearing and mosaic symptoms were sampled and analyzed with small RNA deep sequencing. By denovo assembly the raw sequences of virus-derived small interfering RNAs (vsiRs) and whole genome amplification of malva vein cleaning virus SX strain (MVCV-SX) by specific primers targeting identified contig gaps, the full-length genome sequences (9,645 nucleotides) of MVCV-SX were characterized, constituting of an open reading frame that is long enough to encode 3,096 amino acids. Phylogenetic analysis showed that MVCV-SX was clustered with euphorbia ringspot virus and yam mosaic virus. Further analyses of the vsiR profiles revealed that the most abundant MVCV-vsiRs were between 21 and 22 nucleotides in length and a strong bias was found for “A” and “U” at the 5′-terminal residue. The results of polarity assessment indicated that the amount of sense strand was almost equal to that of the antisense strand in MVCV-vsiRs, and the main hot-spot region in MVCV-SX genome was found at cylindrical inclusion. In conclusion, our findings could provide new insights into the RNA silencing-mediated host defence mechanism in A. rosea infected with MVCV-SX, and offer a basis for the prevention and treatment of this virus disease.
Alcea rosea (L.) is a perennial ornamental herb belongs to Althaea genus, Malvaceae family. This plant is native to China and nowadays distributed over many parts of the world, especially in tropical and temperate regions. As a traditional Chinese medicine plant, its flowers, leaves, roots and seeds possess various pharmacological effects, such as anti-inflammation, anti-oxidation (Liu et al., 2014), blood glucose reduction, hemostasis, detoxification (Zhang et al., 2015) and anti-tumor activity (Choi et al., 2014). However, as the planting area of A. rosea continues to expand, the risk of viral infection is becoming more serious, leading to a negative impact on its pharmacodynamic quality and ornamental value. At present, the viruses infected A. rosea mainly include hollyhock leaf crumple virus (HLCrV), zucchini yellow mosaic virus (ZYMV), althaea rosea enation virus (AREV), malva vein clearing virus (MVCV), cotton leaf curl Bangalore virus (CLCGV), and hollyhock yellow vein mosaic virus (HoYVMV) (Abdel-Salam et al., 1998; Bigarré et al., 2010; Choi et al., 2002; Menzel et al., 2010; Srivastava et al., 2014; Venkataravanappa et al., 2013). During 2015-2017, our laboratory research group has found that A. rosea virus disease is common in the growth area in Shanxi province (China). The infected A. rosea plant exhibits typical mottled leaves, yellow veins and leaf-curling symptoms, with an incidence rate of 80% (Yang et al., 2017), which seriously affects its ornamental and economic values. MVCV-infected A. rosea was first discovered in China in 2017.
MVCV first appeared in mallows in Germany, which causes mosaic and pulse symptoms. Subsequently, it was reported in A. rosea in Spain (Lunello et al., 2009), Germany (Menzel et al., 2010), Hungary and Yugoslavia (Horváth et al., 2010), Italy (Parrella et al., 2015), and China (Yang et al., 2017). Therefore, MVCV is considered a worldwide plant viral disease that causes mosaic and vein-clearing symptoms (Hein, 1959). However, up to date, the full length sequence of MVCV has yet to be reported. In the present work, A. rosea leaves with vein-clearing and mosaic symptoms were sampled in Shanxi Province (China), and subjected to deep small RNA sequencing and bioinformatics analysis. Our findings indicated that A. rosea was co-infected by the MVCV strain SX (MVCV-SX), tobacco virus 1 (TV1), and watermelon mosaic virus (WMV). Furtherly, the full-length genome sequences of MVCV-SX was uncovered and MVCV-derived small interfering RNA (MVCV-vsiR) was profiled. And as far as we know, this is the first report of a full-length genomic sequence of MVCV. This study gives a better understanding of the mechanism of RNA interference-mediated antiviral immunity in A. rosea infected with MVCV-SX and provides a detailed basis for the prevention and treatment of plant viral disease.
Materials and Methods
Sample collection.
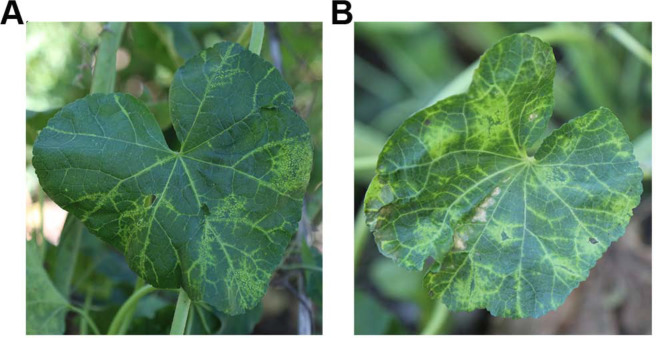
As shown in Fig. 1, A. rosea leaves with vein-clearing and mosaic symptoms were sampled in Taigu, Shanxi province in 2018, and kept in liquid nitrogen until further analysis.
RNA isolation and high-throughput sequencing.
Total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s protocol. The integrity and purity of RNA samples were evaluated with agarose gel electrophoresis and Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), respectively. The qualified RNA samples were immediately transported on dry ice to Biomarker Technologies Co. Ltd. (Beijing, China). The identification and quantification of small RNAs were conducted by small RNA library preparation (TruSeq Small RNA Library Prep Kit, Illumina, San Diego, CA, USA) and next-generation sequencing (HiSeq 4000 System, Illumina).
Bioinformatic analysis of vsiRs.
After sequencing, raw reads were processed for removal of adaptor sequences by using the Perl script. FASTX-toolkit (http://hannonlab. cshl.edu/fastx_toolkit/) was used to trim and filter out low quality reads (score < 20). Small RNAs ranged from 20 to 24 nucleotides were extracted and subsequently analyzed. Velvet was used to assemble the raw reads into contigs (Zerbino and Birney, 2008), which were then searched for vsiRs and their corresponding protein sequences by using BLASTn (sequence similarity > 90% and length coverage > 90%) and BLASTx, respectively. Reference-based mapping/alignment of vsiRs was conducted using the Bowtie software package with no more than three mismatches allowed (Langmead et al., 2009). Small RNA sequences were further evaluated according to a previously reported method (Xia et al., 2014).
Amplification and bioinformatic analysis of MVCV-SX full-length genome sequences.
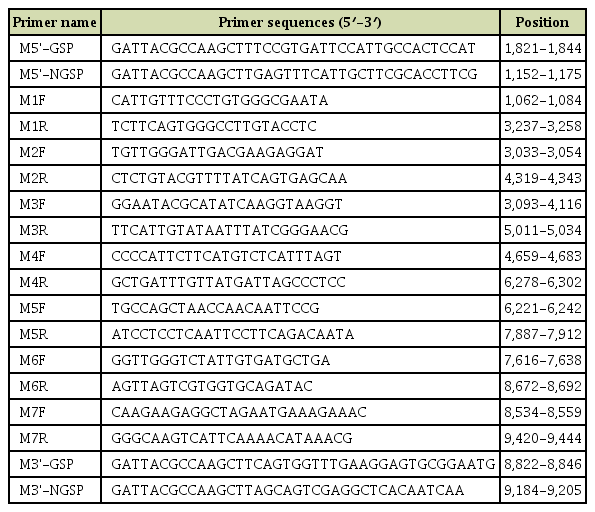
The whole genome sequence data of MVCV were acquired using 5' and 3' rapid amplification of cDNA end (RACE) technology as well as reverse transcription polymerase chain reaction (RT-PCR)–based amplification in order to cover all the missing gaps. The sequences of the seven pairs of specific primers are listed in Table 1.
The first-strand cDNA was obtained from the dsRNA by RT-PCR using Oligo(dT)18 primer and M-MLV reverse transcriptase (Promega, Madison, WI, USA) at 42°C for 1 h followed by 70°C for 15 min. Subsequently, PCR was initiated with Taq DNA polymerase (Sangon Biotech, Shanghai, China) under the previously mentioned reaction conditions (Niu et al., 2018), while RACE was conducted by a RACE kit (TaKaRa) as per the manufacturer’s instructions. Following electrophoresis, the qualified PCR products were purified using a commercial purification kit (Sangon Biotech, Shanghai, China) according to the rec ommendations from manufacturer. The target PCR products were directly ligated with pMD18-T vector (TaKaRa), and then introduced into competent DH5α cells for Sanger DNA sequencing.
Assembly of MVCV-SX sequences was conducted using the SeqMan software in DNASTAR package (Madison, WI, USA). Alignment of multiple amino acid and nucleotide sequences was performed by the DNAMAN software (Lynnon Biosoft, Quebec, Canada). Phylogenetic relationships of MVCV-SX with 89 virus species in the Potyviridae family were determined by the neighbor-joining method using MEGA 7 with the ClustalW algorithm. Bootstrap test (1,000 replicates) was carried out to estimate the significance of each interior branch (Tamura et al., 2007). The putative cleavage sites of different functional proteins in MVCV were evaluated by comparing its own sequence with other virus species (Adams et al., 2005).
Results
De-novo assembly of small RNA reads and characterization of MVCV-SX isolate.
In total, 42,123,237 raw sequences were obtained from the small RNA library. After removing the poor quality reads, 37,017,283 trimmed reads were used for further analyses. De-novo genome assembly with Velvet generated 2651 contigs, in which 15 contigs had a significant similarity to the genomic sequence of MVCV (KX462993.1) by up to 80%, as revealed by the BLASTn analysis. In addition, 41 and 74 contigs shared sequence similarity with TV1 and WMV by up to 65% and 75%, respectively. Seven specific primers targeting identified contig gaps were used to amplify the full-length sequences of MVCV-SX, and the results showed that the entire genome was 9,645 nucleotides in size (GenBank aaccession no. MN116683).
Genome structure analysis of MVCV-SX.
The 5′ and 3'-untranslated terminal regions (UTRs) of MVCV-SX were determined to be 64 and 290 nucleotides in length, respectively, and an open reading frame was identified to encode a large polyprotein (approximately 349.8 kDa) with 3,096 amino acids. Consistent with other potyviruses, this polyprotein could be cleaved into 11 mature proteins (CI, HC-Pro, CP, NIa, NIb, P1, P3, P3N-PIPO, VPg, 6K1, and 6K2), and 9 putative protease cleavage sites were estimated (Fig. 2).
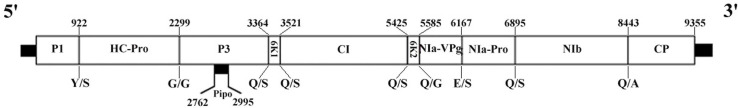
Genome organization of the SX strain of malva vein clearing virus. Open reading frames are denoted by boxes. The number above each box indicates the nucleotide position, while the letter below each box represents the expected cleavage site.
P1 proteinase (nucleotide position: 65-922) is consisted of 286 residues (31.3 kDa), and its cleavage site is located at the carboxyl-terminal end of MEHY/S. P1 appears to be less conserved, as indicated by the location of start codon in this protein. Helper component-proteinase (HC-Pro; nucleotide position: 923-2,299) is composed of 459 residues (51.7 kDa), and its cleavage site is located at YTVG/G. It contains FRNK motif (residues 468-671) that is associated with the activity of RNA silencing suppressor (Shiboleth et al., 2007). P3 protein (nucleotide position: 2,300-3,364) is comprised of 355 residues (40.7 kDa), and its cleavage site is located at VQHQ/S. A putative P3N-PIPO protein (nucleotide position: 2,762-2,995) is consisted of 78 residues, and a highly conserved GAAAAAA slippage motif is located at the 5'-end of PIPO open reading frame. The 6K1 (nucleotide position: 3,365-3,520) is consisted of 52 residues (5.9 kDa), and its cleavage site is located at VYHQ/S. Cylindrical inclusion (CI; nucleotide position: 3,521-5,425) is comprised of 635 residues (70.5 kDa), and its cleavage site is located at VHHQ/S. CI encompasses both DECH (residues 1,326-1,329) and GSGKSTGLP (residues 1,240-1,248) motifs that belong to a superfamily of helicase-like proteins (Fernández et al., 1997). 6K2 (nucleotide position: 5,426-5,584) is consisted of 53 residues (6.1 kDa), and its cleavage site is located at VLFQ/S. Nuclear inclusion protein a (NIa) has 2 domains, namely, NIa-Pro and NIa-VPg. Viral genome-linked protein (VPg; nucleotide position: 5,585-6,166) spans over 22.2 kDa (194 amino acids), and its cleavage site is located at VEFE/S. VPg can affect the main steps of potyviral infection cycle such as viral movement and multiplication (Rajamäki et al., 2014). Nnuclear inclusion protein-a protease (NIa-Prol nucleotide position: 6167-6895) is responsible for the cleavage of most viral precursor polyproteins (Adams et al., 2005). This protein is consisted of 243 residues (27.5 kDa), and its cleavage site is located at VRVQ/S. It has been reported that NIa-Pro is responsible for the cleavage at 6K1/CI, 6K2/VPg, CI/6K2, P3/6K1, NIa-Pro/NIb, NIb/CP and VPg/NIa-Pro sites. Nuclear inclusion body b (NIb; nucleotide position: 6,896-8,443) is comprised of 516 amino acids (59.1 kDa), and its cleavage site is located at VYHQ/A. NIb encompasses RdRp motif GDD (residues 2,628-2,630). Lastly, CP protein (nucleotide position: 8,444-9,355) contains 304 residues (34.2 kDa), and constitutes both C- and N-terminal domains.
Phylogenetic analysis of MVCV-SX.
To study the evolutionary relationship of MVCV-SX, the phylogenetic tree of the members of genus Potyvirus (denoted by red) and 7 other members of the family Potyviridae (indicated in blue), such as Rymovirus, Brambyvirus, Poacevirus, Tritimovirus, Ipomovirus, Macluravirus, and Bymovirus, was constructed by a neighbor-joining algorithm using MEGA 7 program. As shown in Fig. 3, MVCV-SX was highly associated and clustered with yam mosaic virus (YMV) and euphorbia ringspot virus (EuRSV). Moreover, the amino acid and nucleotide sequences of MVCV-SX were evaluated using the DNAMAN program. Sequence-based alignment of MVCV-SX RNA genome with YMV demonstrated that 54.24% and 50.08% of nucleotide and amino acid sequences, respectively, were identical to each other.

Phylogenetic relationships of malva vein clearing virus with other 82 Potyvirus species and a representative of each Potyviridae genus such as Rymovirus, Brambyvirus, Poacevirus, Tritimovirus, Ipomovirus, Macluravirus and Bymovirus. Note: Potyvirus including: asparagus virus 1 (AIY55493.1), banana bract mosaic virus (ABI34615.1), bean common mosaic virus (AAM60816.1), bean common mosaic necrosis virus (ARI46491.1), bean yellow mosaic virus (NP-612218.1), beet mosaic virus (NP-954611.1), bidens mottle virus (YP-003778191.1), calla lily latent virus (ABO77135.1), carrot thin leaf virus (YP-009091824.1), celery mosaic virus (YP-004376199.1), chilli veinal mottle virus (NP-982308.1), clover yellow vein virus (AQY10116.1), cocksfoot streak virus (NP-620483.1), colombian datura virus (YP-007346986.1), cowpea aphid-borne mosaic virus (AEB34826.1), dasheen mosaic virus (NP-613274.1), endive necrotic mosaic virus (ARF07717.1), freesia mosaic virus (CAR58104.1), gloriosa stripe mosaic virus (ABR88099.1), hippeastrum mosaic virus (YP-006382256.1), hyacinth mosaic virus (ASC55665.1), iris mild mosaic virus (AEV51834.1), iris severe mosaic virus (YP-009224125.1), johnsongrass mosaic virus (NP-619668.1), keunjorong mosaic virus (YP-004934107.1), konjac mosaic virus (YP-529485.1), leek yellow stripe virus (NP-659009.1), lettuce mosaic virus (NP-619667.1), lily mottle virus (BAJ10467.1), maize dwarf mosaic virus (NP-569138.1), moroccan watermelon mosaic virus (YP-001552410.1), narcissus yellow stripe virus (YP-002308453.1), onion yellow dwarf virus (NP-871002.1), ornithogalum mosaic virus (YP-006989380.1), papaya ringspot virus (NP-056758.1), passion fruit woodiness virus (YP-004063671.1), pea seed-brone mosaic virus (CAA62014.1), peanut mottle virus (NP-068348.2), peanut stripe virus (AAX82171.1), pecan mosaic-associated virus (ANH79182.1), pennisetum mosaic virus (AGF29582.1), pepper mottle virus (AAA46903.1), pepper severe mosaic virus (YP-778468.1), pepper veinal mottle virus (YP-002519375.1), peru tomato mosaic virus (NP-787937.1), plum pox virus (NP-040807.1), pokeweed mosaic virus (YP-008719787.1), potato virus A (CAA79766.1), potato virus V (NP-659008.1), potato virus Y (NP-056759.1), scallion mosaic virus (CAC87085.1), shallot yellow stripe virus (YP-331412.1), sorghum mosaic virus (AAB70862.2), soybean mosaic virus (AGR55589.1), sugarcane mosaic virus (AAM18491.1), sweet potato latent virus (YP-007697620.1), tobacco etch virus (AIZ00998.1), tobacco vein mottling virus (NP-056867.1), turnip mosaic virus (AAN08045.2), watermelon mosaic virus (ABD59007.1), white lupin mosaic virus (ABG33691.1), wild potato mosaic virus (NP-741959.1), wisteria vein mosaic virus (ANK58176.1), yam mosaic virus (YP-022751.1), zucchini yellow mosaic virus (NP-477522.1), sweet potato virus 2 (AJS10743.1), sweet potato virus G (AJS10736.1), kalanchoe mosaic virus (APX54983.1), jasmine virus T (APZ75428.1), wild onion symptomless virus (BAV17839.1), japanese yam mosaic virus (AJD23399.1), lily virus A (AEN25474.2), papaya leaf distortion mosaic virus (AMB72891.1), sunflower ring blotch virus (AQU42721.1), chilli ringspot virus (APW85806.1), euphorbia ringspot virus (AOT35732.1), arracacha mottle virus (ABI97034.2); Macluravirus including: artichoke latent virus (AHC72297.1); Bymovirus including: barley yellow mosaic virus (CAA10637.1); Brambyvirus including: blackberry virus Y (AAX87001.1); Rymovirus including: ryegrass mosaic virus (CAA70983.1); Ipomovirus including: sweet potato mild mottle virus(CAA97466.1); Poacevirus including: triticum mosaic virus (ABO41208.2); Tritimovirus including: wheat streak mosaic virus (AAM48237.1)
MVCV-derived small RNA characterization analysis.
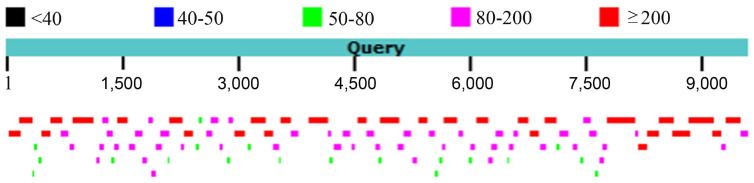
The complete genome sequence of MVCV-SX was obtained by a combination seven specific primers pairs covering all gaps amplification and 5′ and 3′ RACE. The comparison between full-length genome sequences and 2651 assembled contigs showed that 97 contigs were found to be aligned with MVCV-SX, and the distribution of these contigs were almost completely covered in the MVCV-SX genome (Fig. 4). The longest contig was 256 nucleotides, and the 97 contigs (8,800 nucleotides) could span over 92% of the full-length genome sequences.
Previous studies have suggested that DCL proteins play an important role in RNA silencing mechanism for antiviral defenses in plants, where DCL4 generates the most abundant 21 nucleotide vsiRs and DCL2 produces 22 nucleotide vsiRs (Bologna and Voinnet, 2014; Garcia-Ruiz et al., 2010; Llave, 2010; Zhang et al., 2012). The size class analysis of MVCV-vsiRs revealed that vsiRs with 21-22 nucleotide size were the most abundant (Fig. 5A). Among them, the 21 nucleotide size was the most common (68.6%), followed by the 22 nucleotide size (20.9%). Therefore, we speculate that DCL2 and DCL4 both play a crucial role in MVCV-vsiR replication.
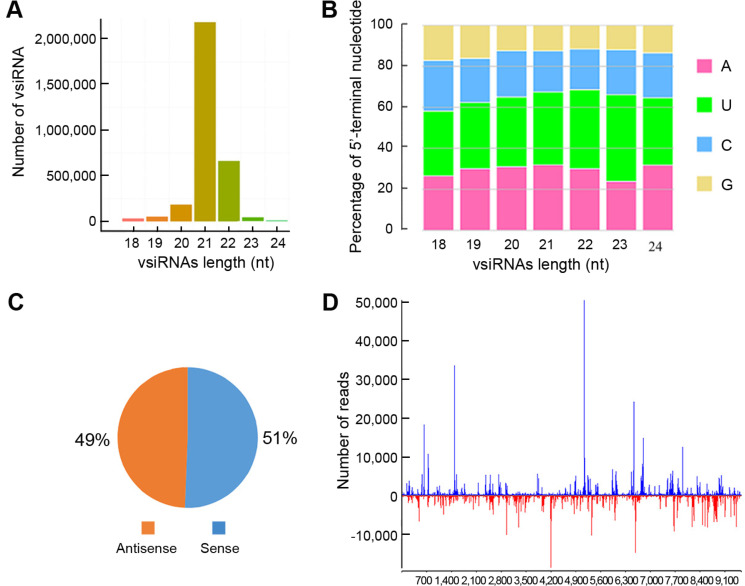
Characterization of malva vein clearing virus strain SX (MVCV-SX) virus derived small interfering RNAs (vsiRs). (A) Length distributions of MVCV-vsiRs in the infected Althaea rosea. (B) Relative frequencies of 5′-terminal sequences in the 18-24 nucleotide MVCV-vsiRs. (C) Strand-specific polarity of MVCV-vsiRs assigned to sense-antisense gene pairs. (D) Hot-spot locations of the MVCV-vsiRs. The horizontal axis indicates the relative position in MVCV-SX genome, while the vertical axis denotes the number of MVCV-vsiRs. The bars on the top and bottom of the axes denote the reads of vsiRs produced by the sense and antisense strands, respectively.
Several reports have shown that vsiRs are recruited by diverse Argonaute (AGO) proteins, which rely primarily on their 5′-terminal sequences (Mi et al., 2008; Takeda et al., 2008). To further examine the possible relationships between vsiRs and various AGO proteins, the relative frequencies of the 5′-terminal sequences of vsiRs were determined. Our findings demonstrated that the nucleotide "A" and "U" were more abundant than "C" and "G" at the 5′-termial end of the 21 nucleotide vsiRs. Furthermore, the proportions of the 5'-terminal starting nucleotide bases starting with "A", "U", "C" and "G" were 31.96%, 35.47%, 19.98% and 12.58%, respectively (Fig. 5B), suggesting the preferable existence of "A" and "U" at the 5′-terminal sequences of MVCV-vsiRs.
Polarity assessment revealed that MVCV-vsiRs constituted 51% and 49% of the sense and antisense strands of MVCV-SX genome, respectively (Fig. 5C). The ratio was determined to be 1:1, suggesting that MVCV-vsiRs are generated from the dsRNA replicative intermediates of MVCV-SX.
To further investigate into their origins, the vsiRs produced by both sense/antisense strands were mapped to the MVCV-SX genome (Fig. 5D). Our data showed a heterogeneous distribution of vsiRs along the sense and antisense strands of MVCV-SX genome. Two large hotspots were detected at 1,500-1,800 nucleotide and 5,100-5,400 nucleotide sites in the virus sense strand. These two hotspots mainly corresponded to the coding regions of HC-Pro and CI in MVCV-SX genome, especially for CI coding region, and the total amount of siRNA reached 50,000. These findings suggest that the coding regions are associated with the secondary structure of vsiRs, and a large amount of vsiRs may be inclined to comply in these regions.
Discussion
MVCV is a member of the genus Potyvirus, which has been first reported in Germany in 1957. It was subsequently found that the host of MVCV was restricted to Malvaceae plants, and most of them were Althaea genus (Horváth et al., 2010). In 2009, Lunello et al. amplified a 1,011 nucleotide MVCV genomic fragment using the universal primer of Potato Virus Y (PVY) virus, including the artial NIb gene, CP gene and 3′ UTR. In 2015, Parrella et al. amplified a total of 1,832 nucleotides of NIb, CP gene and 3′ UTR, and showed a low geographical variation in CP gene among two Italian isolates, six Spanish isolates and one Mexican isolate. In 2017, Yang et al. detected MVCV on hollyhock by sequence-independent amplification and amplified CP gene, which represents the first report of MVCV in China. However, the full-length sequences of MVCV have not been obtained due to the low titer of the virus. In this work, the existence of MVCV was detected on the leaves of A. rosea showing the symptoms of mosaics and veins via next-generation sequencing, and the first complete genome of MVCV-SX was obtained by selfassembly of vsiRs. Furthermore, the vsiR profiles derived from MVCV-SX were further characterized, which could provide new insights into the RNA silencing-mediated host defense against MVCV-SX and guide the design of RNAibased strategies for antiviral treatments.
RNA silencing is a basic antiviral defense mechanism by multi-functional proteins, including AGO and DCL (Bologna and Voinnet, 2014; Llave, 2010). Several studies (Bouché et al., 2006; Deleris et al., 2006; Garcia-Ruiz et al., 2010; Zhang et al., 2012) have indicated that DCL2 plays a key role in vsiR replication and generates the most abundant 22 nucleotide vsiRs, while DCL4 produces 21 nucleotide vsiRs in virus-infected plants. In this work, the size-class analysis of MVCV-vsiRs showed that the 21 and 22 nucleotide sizes were the most prominent, which account for 89.5% of total MVCV-vsiRs in MVCV-SX-infected plants. Such sizes were similar to those of vsiRs in plants during the prevention of virus damage, indicating that these vsiRs are primarily cleaved by DCL2 and DCL4. Therefore, it can be speculated that DCL2 and DCL4 play a key role in host defense against MVCV. Besides, several AGO proteins have been shown to be associated with host immune responses in Arabidopsis thaliana, which can be driven by the 5′-terminal sequence. For instance, AGO1 is predominantly related to small RNAs with a 5′-U, while AGO2 and AGO4 are more to 5′-A (Mi et al., 2008; Takeda et al., 2008). The MVCV-vsiRs of 21 and 22 nucleotides in length start with the bases "A" and "U", implying that these vsiRs are potentially recruited by AGO1, AGO2 or AGO4. However, in the long-term struggle between the study of A. rosea and MVCV, it remains unclear whether AGO protein has such preference. Thus, further research is needed to verify the roles of DCL2 and DCL4 as well as AGO1, AGO2 and AGO4 proteins in antiviral defense during the interaction between MVCV-SX and A. rosea plants.
Acknowledgments
This study was supported by National Natural Science Foundation of China (No. 31772130), China Agriculture Research System (No. CARS-21) and Modern Agro-industry Technology Research System (No. 2020-05).