Insights into the Incidence of Watermelon chlorotic stunt virus Causing Yellowing Disease of Watermelon in Western and Southwestern Regions of Saudi Arabia
Article information
Abstract
During the spring season of 2014, a total of 148 melon and watermelon leaf samples were collected from symptomatic and asymptomatic plants in the western and southwestern regions of Saudi Arabia and were tested for the presence of Watermelon chlorotic stunt virus (WmCSV) and other suspected cucurbit viruses by double antibody sandwich enzyme-linked immunosorbent assays. Ninety-eight samples were found to be positive for the presence of WmCSV, nine samples were positive for the presence of Cucurbit yellow stunting disorder virus (CYSDV), and 22 showed a mixed infection with both WmCSV and CYSDV. No other cucurbit viruses were detected in any of the samples. Host range experiments revealed that eight out of fourteen tested plant species were susceptible to WmCSV. PCR products of approximately 1.2 kb were obtained after amplification using primers specifically targeting the coat protein region of WmCSV. Positive PCR results were confirmed by dot blot hybridization. Coat protein gene sequences from eleven WmCSV isolates indicated that the highest identity was between the 104WMA-SA isolate from the Wadi Baish location and a previously reported isolate from the AL-Lith location in Saudi Arabia. The lowest identity was observed between the 42WMA-SA isolate and an isolate from Palestine.
Introduction
The cultivated species of the gourd family (Cucurbitaceae) are collectively known as cucurbits (Wiley and Sons, 2012). They are native to the subtropical and tropical Americas, Africa, and Asia (Robinson and Decker-Walters, 1997). Among them, watermelon (Citrullus lanatus L.) and melon (Cucumis melo L.) are valuable cash crops grown in temperate and tropical regions worldwide (Wang et al., 1997; Erickson et al., 2005). Both watermelon and melon crops have been reported to be affected by many plant viruses, including Cucumber mosaic virus (CMV), Cucumber green mottle mosaic virus (CGMMV), Watermelon mosaic virus (WMV), Zucchini yellow mosaic virus (ZYMV), Squash mosaic virus (SqMV), Cucurbit yellow stunting disorder virus (CYSDV), and Watermelon chlorotic stunt virus (WmCSV). These viruses can adversely affect the durability, quality, and yield of the crop (Zitter et al., 1996).
WmCSV is a bipartite member of the genus Begomovirus. It is transmitted in a circulative persistent manner by Bemisia tabaci, which is able to parasitize a variety of hosts worldwide (Bedford et al., 1994a; Bedford et al., 1994b). This virus was first identified in Yemen (Jones et al., 1988; Walkey et al., 1990; Bedford et al., 1994b) and it infects watermelon (C. lanatus L.), cucumber (Cucumis sativus L.), squash (Cucurbita pepo L.), and melon (C. melo L.; Bananej et al., 2002; Kheyr-Pour et al., 2000; Zerbini et al., 2017). In 2014, yellowing disease symptoms were observed in many fields of watermelon and melon in the western and southwestern regions of Saudi Arabia. A careful inspection of symptoms found that they were similar to those previously described (Al-Saleh et al., 2014; Bananej et al., 2002) to be caused by WmCSV, indicating a possible involvement of this virus. Since this virus is an important pathogen that infects cucurbits and produces yellowing disease, its etiology needs to be investigated and its presence needs to be confirmed. The objective of this investigation was to serologically, biologically, and molecularly identify the WmCSV causing yellowing disease of watermelon and melon in western and southwestern regions of Saudi Arabia, and to determine the phylogenetic relationship between Saudi WmCSV isolates and other isolates available in Gen-Bank.
Materials and Methods
Source of virus isolates and serological and biological identification
During the spring seasons of 2014 and 2015, a total of 148 samples (139 from watermelon, five from wild watermelon, four from melon) were collected from naturally symptomatic plants showing virus-like symptoms, including yellowing, mosaic, mottling, and stunting (Fig. 1). Samples were collected from the western (Jeddah, Al-Lith, Tofeel, Asfan, and Ghonfada) and southwestern (Jezan, Wadi Baish, and Abu Arish) regions of Saudi Arabia. All samples were wrapped in plastic bags and preserved at 4°C or −80°C for short- and long-term storage, respectively (Nelson and Bushe, 2006).

Yellowing, mosaic, and mottling symptoms observed in watermelon plants in the field after infection with WmCSV.
Commercial ELISA kits (AC Diagnostic, Fayetteville, Arkansas, USA) were used, as described by Clark and Adams (1977), for the detection of cucurbit viruses infecting watermelon and melon, including CMV, CGMMV, WMV, ZYMV, SqMV, CYSDV, and WmCSV. A WmCSV-positive double antibody sandwich enzyme-linked immunosorbent assays (DAS-ELISA).
Sample from Al-Lith (29WMA-SA) was used as a source of inoculum and propagated into the propagative host, C. lanatus, using the whitefly (Bemisia tabaci) as a vector. A total of 14 plant species (C. lanatus, C. melo, Cucurbita maxima, Cucurbita pepo, Cucumis sativus, N. tabacum, Chenopodium quinoa, C. album, Amaranthus viridis, Lagenaria siceraria, Momordica charantia, Vigna radiata, Solanum nigrum, and Datura stramonium) representing several families were tested in the host range experiment. Two sets, each having five plants representing each species and including a control plant, were used. All inoculated host plants were tested by PCR and/or dot blot hybridization to confirm the presence of the WmCSV.
Non-viruliferous colonies of B. tabaci were raised on squash (C. pepo) seedlings grown in muslin-covered (insect-proof) cages. A group of 50 non-viruliferous B. tabaci was allowed to access WmCSV-infected C. lanatus plants for an acquisition access period of 48 h. After completion of the acquisition period, ten viruliferous whiteflies were transferred to healthy host range plants at the three-leaf stage and were allowed to feed for a 48-h inoculation access period (IAP). The non-viruliferous whiteflies were allowed to feed on control plants for a similar period of time. Following the IAP, whiteflies were eradicated by treating plants with insecticide (acetamiprid), and plants were then kept in insect-proof cages under controlled conditions and monitored for the development of symptoms for up to one month (Al-Musa et al., 2011). A description of the symptoms that developed in each species was recorded. All inoculated and control plants were tested by PCR and/or dot blot hybridization to confirm the presence of the virus. The experiment was repeated to confirm the results of the first inoculation.
PCR identification
For WmCSV identification, total nucleic acids were extracted from 26 watermelon samples tested by DAS-ELISA, five melons, and four wild watermelons, as described by Dellaporta et al. (1983). PCR was performed using primers specific for the coat protein (CP) region of the WmCSV genome (WmA150F 5′-gtc agt atg tgg gat cca ttg c-3′ and WmA1350R 5′-gca aat acg att caa cca caa cc-3′), using 2X KAPA Taq Ready Mix (KAPA Biosystems, Boston, Massachusetts, USA). PCR was performed using a thermal cycler (Eppendorf, AG, Hamburg, Germany), according to the method described by Ali-Shtayeh et al. (2014). DNA was visualized, as described by Sambrook and Russell (2001), and photographed using a DNA documentation gel analysis system (IN GENIUS, Syngene Bio Imaging, Cambridge, UK). A DNA Ladder (100 bp RTU; Genedirex, Inc. Taoyuan, Taiwan) was used to determine the size of the amplified PCR products.
Molecular hybridization for the detection of WmCSV
A WmCSV-specific probe was synthesized by PCR amplification using a PCR DIG Probe Synthesis Kit (Roche Diagnostics, GmbH, Germany). All 148 samples were prepared for dot blotting on nitrocellulose membrane according to the methods of Pallas et al. (1998) and Podleckis et al. (1993). Prehybridization, hybridization, and colorimetric detection were performed in a hybridization oven (Amersham Biosciences, Piscataway, NJ, USA) following the protocol recommended by Boehringer Mannheim, Roche Diagnostics GmbH, Mannheim, Germany. Results were documented by photographing the wet filter.
Nucleotide sequence and phylogenetic analysis for WmCSV-CP
Eleven amplified PCR products of the WmCSV CP gene, representing all the studied locations, were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Central Avenue, Union City, USA), according to the manufacturer’s instructions, and sequenced in both directions at BGI Tech Solutions (Hong Kong, China). The nucleotide sequences of the WmCSV CP gene from Saudi isolates were subjected to the BLASTN algorithm and compared with 21 sequences retrieved from GenBank. Multiple sequence alignments and phylogenetic relationships for these isolates (Table 1) were analyzed and reconstructed using Lasergene V5-05 Software (DNASTAR), Madison, Wisconsin USA.
Results
Serological and biological identification of virus isolates
DAS-ELISA results revealed that 98 out of 139 watermelon samples (70.5%) were infected with WmCSV, nine (6.4%) were infected with CYSDV, and 22 (15.8%) had a mixed infection of both viruses (Table 1). None of the melon or wild watermelon samples were found to be positive for the presence of WmCSV or CYSDV. None of the collected samples from any host showed a positive reaction to other tested cucurbit viruses (CMV, CGMMV, WMV, ZYMV, and SqMV). Results of the host range experiment showed that, out of 14 inoculated plant species, eight were successfully infected with WmCSV. These plant species showed varying symptoms: chlorotic mottling and vein yellowing on C. lanatus; interveinal chlorotic patches on C. melo; interveinal chlorotic patches and chlorotic lesions on C. pepo; interveinal chlorotic patches and mottling on L. siceraria; mosaic on C. maxima, interveinal chlorosis on V. radiate and D. stramonium; mild interveinal chlorotic patches on A. viridis, and no symptoms on C. sativus, M. charantia, N. tabacum, C. quinoa, C. album, and S. nigrum (Fig. 2 and Table 2).
Symptoms observed, PCR, and dot blot hybridization confirmation on some of the host range plants inoculated (left: A, B, C, D, E, F and G) and healthy control plant (right: a, b, c, d, e, f, and g) with the Saudi isolate of WmCSV. (A) chlorotic mottling and vein yellowing on C. lanatus. (B) Interveinal chlorotic patches on C. melo. (C) Symptoms of mosaic on C. maxima. (D) interveinal chlorotic patches and chlorotic lesions on C. pepo. (E) interveinal chlorotic patches and mottling on L. siceraria. (F) interveinal chlorosis on V. radiata. (G) interveinal chlorosis on Datura stramonium. (H) mild interveinal chlorotic patches on A. viridis.
PCR identification
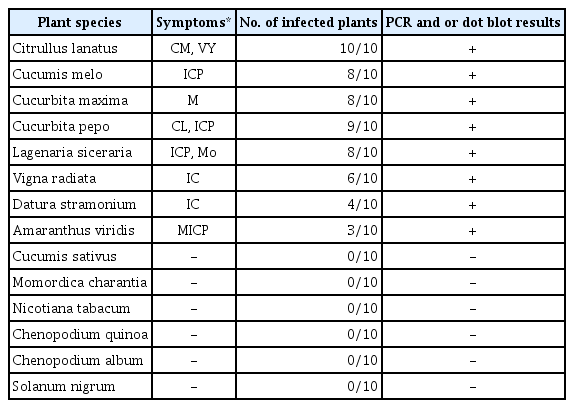
PCR detection, using primers specific for the WmCSV CP gene, positively confirmed WmCSV infection in 26 selected watermelon plant samples. Fig. 3 shows the results of the watermelon samples collected from different locations. A PCR product of 1201 bp was visualized on a 1% agarose gel. No PCR product was amplified from uninfected watermelon (negative control), melon, or wild melon.
Agarose gel (1%) electrophoresis analysis of PCR products for WmCSV (1201 bp) using specific primers for the coat protein gene from selected symptomatic watermelon leaves collected from different locations. Lanes 1–5: from Al-Lith, lanes 6–9: from Wadi Baish, lanes 10–13: from Jeddah, lanes 14–17: from Tofeel, lanes 18–20: from Abu Arish, lanes 21–22: from Asfan, lanes 23–24: from Ghonfada, and lanes 25–26: from Jezan. No PCR amplification was observed in uninfected sample tissue (lane N), melon (lane 27), or wild watermelon (lane 28). Lane L: 100-bp RTU DNA Ladder (Genedirex).
Molecular hybridization for detection of WmCSV
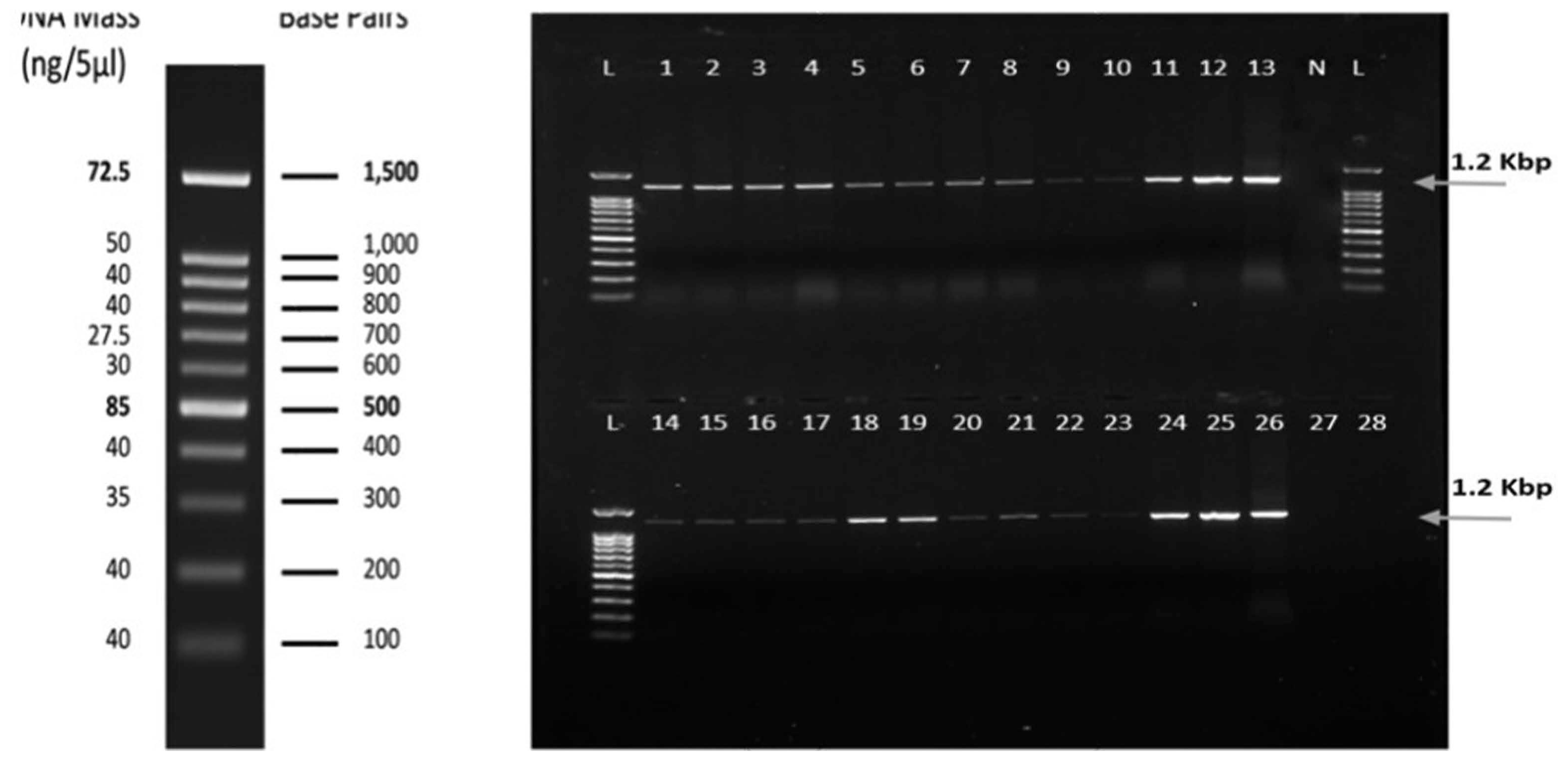
Out of the 148 samples tested using a molecular hybridization assay against WmCSV, 127 were found to be infected with WmCSV. Fig. 4 shows the results of the molecular hybridization assay of some selected watermelon and melon samples collected from different locations. These results confirmed that none of the melon or wild watermelon samples were positive for the presence of WmCSV.
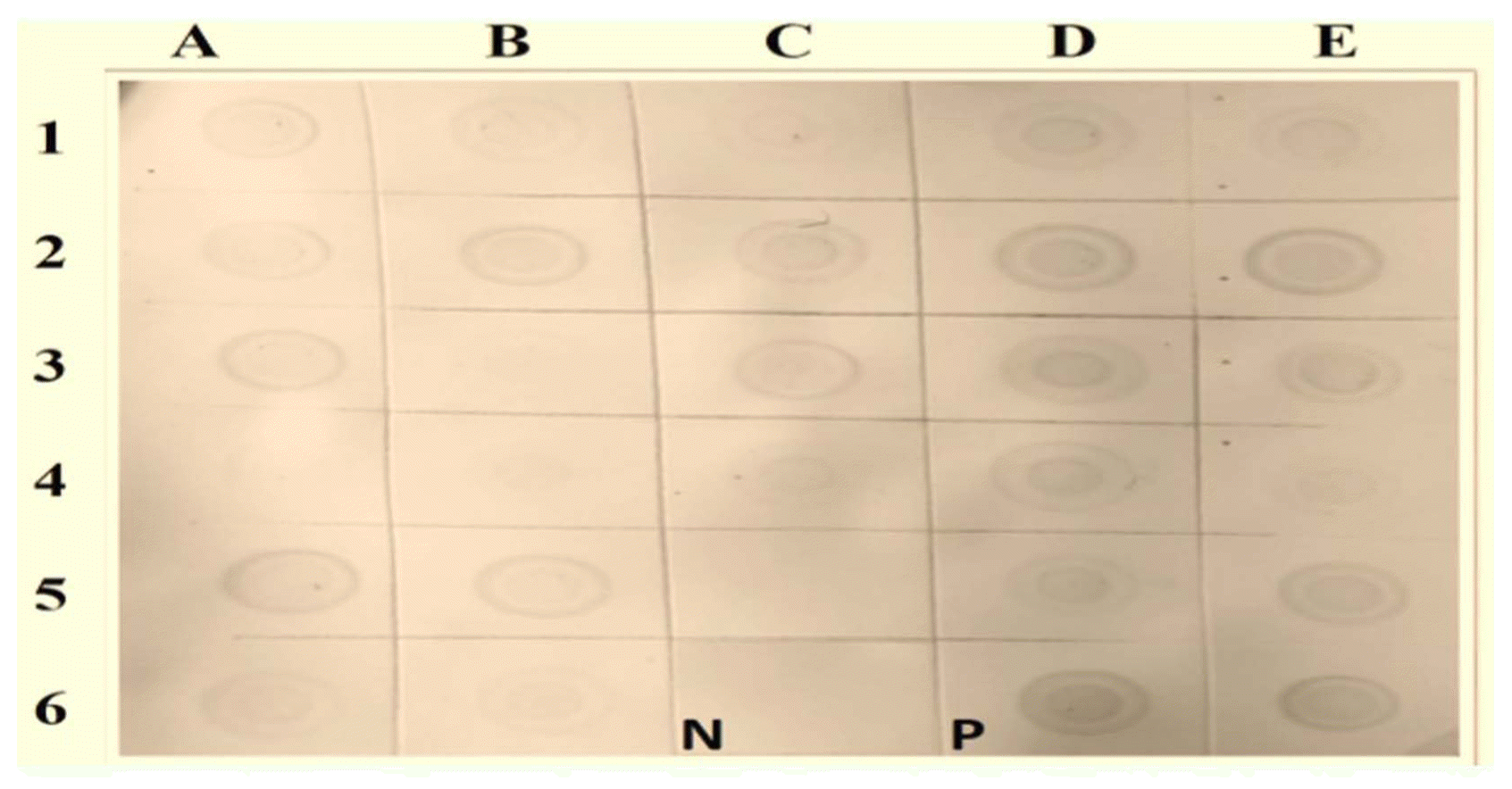
Dot blot hybridization of selected samples using a WmCSV-DIG-DNA probe. Nitrocellulose membranes indicating positive (purple) and negative results. Column A1, 2, 3: from Wadi Baish; Column A 4, 5, 6: from Jeddah; Column B 1, 2, 3: from Jezan; Column B 4, 5, 6: from Asfan; Column C 1, 2, 3: from Ghonfada; Column C 4, 5: from Tofeel; Column D 1, 2, 3, 4, 5: from Abu-Arish; and Column E 1, 2, 3, 4, 5, 6: from Al-Lith. PCR product was used as a positive control (P). No hybridization reaction was observed with uninfected watermelon samples (N).
Nucleotide sequence and phylogenetic analysis of WmCSV-CP genes
Complete sequences of the CP genes from 11 Saudi Arabian WmCSV isolates were determined and submitted to GenBank under the accession numbers given in Table 3 and Table 4. These 32 complete CP gene nucleotide sequences, including those of the Saudi Arabian WmCSV isolates, were analyzed phylogenetically (Fig. 5) using DNASTAR software. All isolates fell within four distinct groups. Group A of the phylogenetic tree contains 17 isolates, with two isolates from Jordan (watermelon and wild mustard (Sinapis arvensis), four from Oman (squash), seven from Palestine (watermelon), and one each from Lebanon (melon), Israel (watermelon), Sudan (watermelon), and Iran (watermelon). Group B contains 13 isolates from Saudi Arabia isolated from watermelon. These isolates include 11 from the present study and 2 previously published Saudi isolates (KM066100 and KJ939448) from the Al-Lith region (Al-Saleh et al., 2014). In contrast, group C and group D each contain a single isolate from Yemen and Sudan isolated from watermelon and Datura innoxia, respectively.

Percentage identity, based on CP sequences between WmCSV Saudi isolates after aligning using the cluster W method
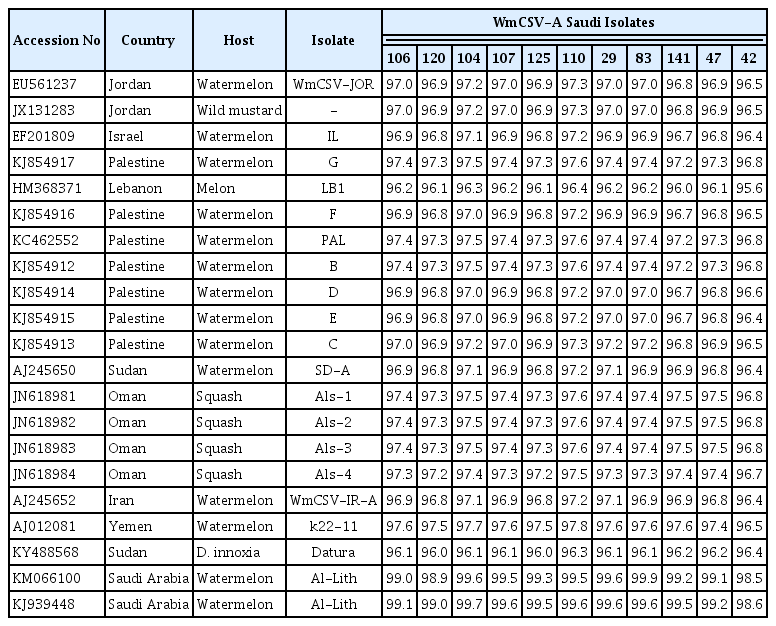
Percentage identity, based on CP sequences of 11 WmCSV Saudi isolates from the present study and 21 isolates available on GenBank, after aligning using the cluster W method
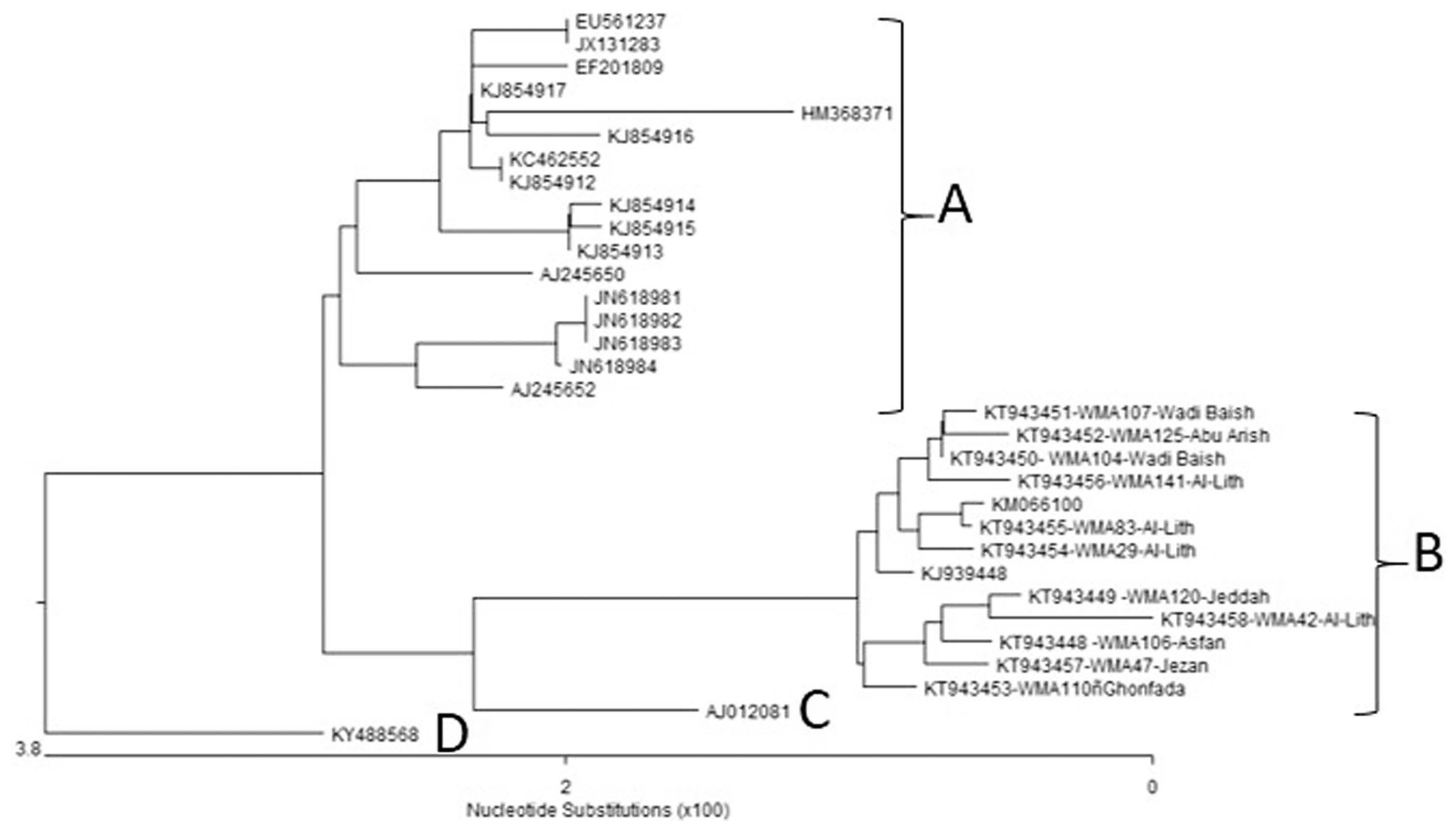
Phylogenetic tree, based on complete coat protein gene sequences of 32 isolates, shows four groups. Group A has all 17 isolates whose sequences are available in GenBank, while group B has 13 Saudi Arabia (11 new and 2 previously known) isolates. Group C and D each contain a single isolate from Yemen and Sudan, respectively.
The nucleotide sequence identity among the 11 Saudi Arabian isolates ranged from 98.6% to 99.9%. The highest identity (99.9%) was found between 104WMA-SA (KT943450) and 107WMA-SA (KT943451) isolated from watermelon from the Wadi Baish location. The lowest identity (98.6%) was found between 29WMA-SA (KT943454), 83WMA-SA (KT943455), and 141WMA-SA (KT943456) isolates from Al-Lith and the 42WMA-SA (KT943458) isolate from Tofeel (Table 3). The nucleotide sequence identities between all Saudi Arabian isolates and other isolates from GenBank ranged from 95.6 to 99.0%. The highest identity (99.9%) was found between the 83WMA-SA (KT943455) WmCSV isolate from the Al-Lith location and a previously reported isolate (KM066100) from the Al-Lith location in Saudi Arabia. The lowest identity (95.6%) was found between the WmCSV KSA isolate, 42WMA-SA (KT943458), and isolate HM368371 from Lebanon (Table 4).
Discussion
WmCSV is considered an important virus infecting cucurbits in many Middle Eastern countries (Abou-Jawdah et al., 2010; Al-Musa et al., 2011; Bananej et al., 2002; Kheyrpour et al., 2000). In the early 2000s, WmCSV was introduced into the Eastern Mediterranean Basin. This virus has been emerging in parallel in Egypt, Israel, Jordan, Lebanon and Palestine over the last decade. Pathogens frequently migrate between Middle Eastern, Arabian Peninsula and North African regions. Seeds and vegetative plant material also move across these national borders and this is known to contribute to the spread of plant viruses. Another reason for the spread of the virus may be the transmission of WmCSV due to uncontrolled migration of whitefly vectors among neighboring countries, thus decreasing the genetic diversity of the virus.
In a scenario where Saudi Arabia has strong agricultural trading ties with neighboring Arab countries including Palestinian, Lebanon, Yemen, Oman, Jordan, and Sudan, WmCSV may arise as a threat to watermelon and other cucurbit crops grown in Arab countries and the rest of the world.
Outside the Red Sea region, the virus has been found in Iran (Bananej et al., 2002; Kheyr-Pour et al., 2000) and between the years 2010–2012, it was reported in Israel, Jordan, Lebanon, and Oman (Abou-Jawdah et al., 2010; Abudy et al., 2010; Al-Musa et al., 2011; Khan et al., 2012). Recently, the virus was detected for the first time in Saudi Arabia (Al-Saleh et al., 2014).
The present study was planned after observing virus-like symptoms, such as yellowing, mosaic, mottling, and stunting in watermelon and melon crops during a field visit to Al-Lith in spring, 2014. During the field survey, along with virus-like symptoms, large populations of whiteflies were also observed. Since WmCSV is transmitted by whiteflies, this may explain the high incidence of disease in the surveyed fields. Disease incidence in some fields was observed to be more than 90%, which is similar to a previous study where disease incidence reached 98% in some watermelon fields surveyed in Palestine (Ali-Shtayeh et al., 2014).
DAS-ELISA results showed that only 19 samples (12.83%) were negative for WmCSV, which is not surprising given that disease incidence was 90% and symptoms were severe.
The high incidence of disease may also be attributed to infection with other viruses and/or nutritional deficiency. However, the majority of samples were infected with WmCSV, so we can confidently assume that WmCSV had a major role in symptom development and disease progression in these areas. There were significantly fewer samples with mixed infection compared to WmCSV infection in this study, whereas previous studies have reported that mixed infections with two or three whitefly-transmitted viruses are common in cucurbit fields in Lebanon, Palestine, and Jordan (Abou-Jawdah et al., 2010; Al-Musa et al., 2011; Ali-Shtayeh et al., 2014). Hence, these viruses pose a great threat to cucurbit crops in Saudi Arabia.
Host range results showed that most of the tested species were susceptible, which indicates that WmCSV infects most members of Cucurbitaceae. Interestingly, cucumbers were found to be negative, although these have been shown to be positive for WmCSV infection in previous studies (Abudy et al., 2010; Al-Musa et al., 2011). However, this result is supported by a previous study by Bananej et al. (2002) in which WmCSV also failed to infect cucumbers.
One possible explanation is that the cucumber variety (Beit alpha) used in our host range experiment was resistant or immune to WmCSV. On the other hand, L. siceraria (bottle gourd) is a newly identified experimental host of WmCSV, as it was found to be a non-host for this virus in previous studies (Abudy et al., 2010; Al-Musa et al., 2011). Other possible explanations for these conflicting results are differences between the virus isolates or even the host preference of the vector populations used. These data show that WmCSV is expanding its host range among members of the Cucurbitaceae family. In addition, A. viridis, a common weed plant, was confirmed to be susceptible to WmCSV for the first time. The susceptibility of A. viridis and D. stramonium to WmCSV may explain how the virus survives during the off-season. Further studies are needed to elucidate these aspects.
Over the last decade, the sensitivity of plant virus detection in a great number of virus-host combinations has improved due to the use of nucleic acid-based techniques instead of serological techniques. Indeed, such techniques are now considered reliable, fast, and inexpensive. The dot blot hybridization technique has been successfully used for the specific detection of WmCSV (Bananej et al., 2002; Kheyr-Pour et al., 2000). In the present study, out of 139 samples, 127 samples (91.3%) were found to be infected, while 12 samples (8.6%) were found to be negative.
Dot blot hybridization results not only confirmed the presence of WmCSV in field samples testing positive with DAS-ELISA, but also in some DAS-ELISA negative samples. This indicates that dot blot hybridization is a more sensitive technique than DAS-ELISA, since it is based on a DNA probe (Sanchez-Navarro et al., 1998).
The presence of PCR products (1201 bp) in all tested samples confirmed the presence of WmCSV in these samples. In the case of WmCSV, the identity of viral isolates was confirmed by nucleotide sequence analysis of the CP gene. Eleven Saudi Arabian isolates had a high nucleotide sequence identity percentage among themselves, ranging from 98.6% to 99.9%. All isolates sharing the same group, A2, along with two other isolates from GenBank, which were also from Al-Lith, Saudi Arabia, have a common host i.e., watermelon. This shows that Saudi isolates of WmCSV from the same host are highly similar to each other. Our isolates also show a high identity percentage with isolates from the GenBank database, with an identity range of 96.1% to 99.7% (Table 4). Nucleotide sequence identities (Table 4) show that all but one of the new Saudi isolates are > 97.4% identical to the prior Yemeni isolate and appear to be in a closer relationship with this isolate than the other previously characterized isolates. In addition, the phylogenetic tree shown in Fig. 5 clearly indicates that all Saudi isolates are derived from the same ancestor as the Yemeni isolate. The earlier reported occurrence in Yemen (Jones et al., 1988) also indicates that this may be the source, with an increase in sequence diversity over subsequent years.
WmCSV may have recently entered Saudi Arabia or it may have remained unidentified for a long time due to a lack of research on this virus. The percentage identity between Saudi isolates and other isolates reported in Gen-Bank ranged from 96.6–97.6%. In a similar study, Khan et al. (2012) found that five of the WmCSV isolates from squash that they sequenced shared 99–100% nucleotide sequence identity. Comparisons to sequences available in the databases showed that the WmCSV isolates in this study had a relatively high identity ranging from 95.6.5–99.9.8%. The highest levels of identity were found to be with an isolate of WmCSV originating from Iran (AJ245652). Ali-Shtayeh et al. (2014) conducted a similar study, showing that the isolates they sequenced shared a high identity percentage, ranging from 99–100%, while database comparisons revealed high degrees of sequence identity (i.e., > 93%) with other WmCSV isolates. Al-Musa et al. (2011) also found high degrees of sequence identity (i.e., > 94%) in a database comparison. The existence of WmCSV in the southern region of Saudi Arabia may result in a source of infection for crops grown in the other regions and may also emerge as a recombinant strain in the future. Apart from searching for a source of resistance, control of whiteflies and alternate hosts may be a practical approach to prevent the virus from spreading.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research, King Saud University, Saudi Arabia, for funding this research (Group no. RG-1438-065).