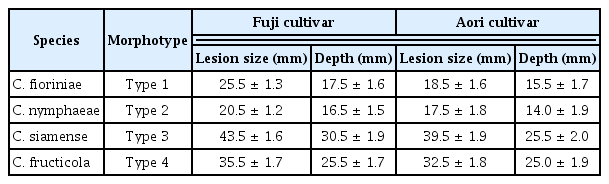
Identification and Characterization of Colletotrichum Species Associated with Bitter Rot Disease of Apple in South Korea
Article information
Abstract
Bitter rot caused by Colletotrichum species is a common fruit rotting disease of apple and one of the economically important disease in worldwide. In 2015 and 2016, distinct symptoms of bitter rot disease were observed in apple orchards in five regions of South Korea. In the present study, infected apples from these regions were utilized to obtain eighteen isolates of Colletotrichum spp. These isolates were identified and characterized according to their morphological characteristics and nucleotide sequence data of internal transcribed spacer regions and glyceraldehyde-3-phosphate-dehydrogenase. Molecular analyses suggested that the isolates of Colletotrichum causing the bitter rot disease in South Korea belong to 4 species: C. siamense; C. fructicola; C. fioriniae and C. nymphaeae. C. siamense and C. fructicola belonged to Musae Clade of C. gloeosporioides complex species while C. fioriniae and C. nymphaeae belonged to the Clade 3 and Clade 2 of C. acutatum complex species, respectively. Additionally, we also found that the isolates of C. gloeosporioides species-complex were more aggressive than those in the C. acutatum species complex via pathogenicity tests. Taken together, our results suggest that accurate identification of Colletotrichum spp. within each species complex is required for management of bitter rot disease on apple fruit in South Korea.
Body
The apple tree (Malus pumila, commonly known as Malus domestica) is a deciduous tree belonging to the family Rosaceae, and apples are the third most traded fruit internationally followed by bananas and grapes (Lynch, 2010). Apple cultivation originated in Central Asia and with time has spread worldwide. Apple is best known for their sweet taste and is extremely rich in important dietary fibers, antioxidants, and flavonoids. The antioxidants and flavonoids in apples help in reducing the risk of developing hypertension, heart disease, cancer and diabetes (Boyer and Liu, 2004). In South Korea, apple is grown in a large amount with the expected cultivation area of 33,600 ha in 2017/2018 and has played a critical role in Korean economic development (Choi and Hinkle, 2017). The major varieties of apples grown in Korea are Tsugaru (Aori) (harvested in August), Hongro (harvested in September), Fuji (harvested from October to November), Yang Kwang (harvested from September to October), and Kam Hong (harvested in October) (Choi and Hinkle, 2017). However, numerous destructive pathogens infect apple trees and cause significant yield loss in apple production worldwide (Munir et al., 2016; Sutton, 1990).
Among these diseases, bitter rot caused by Colletotrichum species is one of the important apple fruit diseases in worldwide and causes significant yield losses to the apple producers (Munir et al., 2016; Uhm, 2010). The symptom of apple bitter rot initially appears as a small circular lesion on fruits that are light to dark brown in color. On mature fruits, these areas may be surrounded by a red halo (Beever et al., 1995; Struble et al., 1950). Under favorable conditions, these lesions subsequently enlarge rapidly and cover the entire fruit surface. As the spots enlarge, the fruit rot symptoms can vary depending on the fungal isolate and the type of spore that initiates infection. Spots initiated by isolates that only produce conidia are sunken and circular with concentric rings (Beever et al., 1995; Struble et al., 1950). Within the rings, copious amounts of conidia are produced in fruiting bodies called acervuli. Under moist, humid conditions, the spore masses appear creamy and are salmon to pink in color. Spots initiated by isolates that produce conidia and ascospores are usually not sunken and are darker brown in color than spots produced by conidial isolates. Within the spots, structures that contain ascospores (perithecia) form in black clumps over the surface (Beever et al., 1995; Struble et al., 1950). As spots enlarge, the rot progresses inward towards the core. After making a cross-section of the infected fruit a V-shaped, brown watery lesion can be observed (Velho, 2014). This is a key diagnostic feature of bitter rot, which distinguishes it from other fruit rots disease. Moreover, the bitter rot pathogens over-winter as perithecia and acervuli in mummified apples, and as mycelia in colonized dead wood, unpruned branches and cuttings that are left on the ground (Sutton, 1990). Without proper disease management, bitter rot can destroy an entire fruit crop in just a few weeks during periods of warm, wet weather (Sutton, 1990).
Genus Colletotrichum is one of the most important pathogenic fungi causing economically significant diseases on the fruits and leaves of a wide range of subtropical, tropical, and temperate crops, fruits, and ornamental plants (Cannon et al., 2012; Sutton, 1992). Moreover, genus Colletotrichum is a cosmopolitan fungal genus and comprises 11 major clades (Jayawardena, 2016). Some Colletotrichum species have wide host ranges, while others can infect only a single host species (Damm et al., 2012; Weir et al., 2012). Diverse species of Colletotrichum can cause bitter rot diseases in apples. A previous report demonstrated that two species of C. gloeosporioides and C. acutatum are the most common species causing bitter rot disease on apple fruit in worldwide (González et al., 2006). In South Korea, C. gloeosporioides and C. acutatum were also recorded as causal pathogens for bitter rot on apples (KSPP, 2009; Lee et al., 2007). Additionally, Park et al. (2018) reported two new species of C. siamense and C. fructicola as causal agents of bitter rot disease of apple in South Korea. However, their identification method using morphological characteristics may not provide reliable characterization because there are no definite morphotaxonomic characters on these species (Bernstein et al., 1995; Cai et al., 2009). Therefore, the objective of this study was to identify Colletotrichum species associated with bitter rot disease of apple in South Korea and to characterize and compare the pathogenicity of the isolated species.
Materials and Methods
Sampling and isolation of the isolated fungus
In 2015 and 2016, symptomatic fruits were collected from some private orchards in different locations of South Korea. Samples of fruits affected by typical disease symptoms were washed with water and dried in a laminar air flow chamber. After surface sterilization, infected fruits tissues were cut into 5 mm pieces and disinfected by dipping in 1% Sodium hypochlorite (NaClO) for 3 minutes, rinsed 3 times with sterile distilled water, and dried on sterilized tissue paper (Nam et al., 2013). After drying, the samples were cultured on 90- mm Petri dishes containing blotter paper and incubated at room temperature (25 ± 2°C) for 12 photoperiods. After 2–3 days, spore layers were picked with a smear loop or autoclaved toothpick and mixed with 1ml of sterilized distilled water supplement with Streptomycin (300 ppm for 1 l) to prevent the bacteria contamination. After mixing with this solution and spore, the mixture was then spread on Water Agar culture medium (containing Streptomycin 300 ppm for 1 l) for 2–3 days at room temperature (25 ± 2°C) (Prihastuti et al., 2009). Two or 3 days after isolation, a single isolated spore of emerging fungi was picked from the sporulating colonies and transferred to new potato dextrose agar (PDA; Difco, Sparks, MD, USA) plates to obtain the pure culture and were further grown for DNA extraction, pathogenicity tests and morphological characterization. The culture plates were checked every day with naked eyes for measuring hyphal growth.
Morphological characterization
To examine the morphological characteristics, fungal species were cultured on PDA and V8 juice media at 25 ± 2°C in the darkness for 7 days. Small discs (5 mm diameter) of mycelium plugs were aseptically punched from actively sporulating areas of 4 – 5 days old culture and transferred to the center of V8 juice and PDA media plate and then incubated at room temperature (25 ± 2°C). Three cultures of every isolates were investigated. After 7 days, colony color, size, texture, and zonation of every culture were recorded from V8 juice and PDA media. Colony colors were named according to the mycological color chart (Rayner, 1970). Colony diameter (mm) was measured and calculated based on the average of the perpendicular diameter and the data were examined to compute the mycelial growth rate (mmd−1). The sporulating pattern was analyzed under a Olympus BX40 dissecting microscope (Olympus optical Co., Ltd, Japan). Conidia of every isolate were characterized using conidia from 10 days old colonies on PDA media, mounted in a drop of lactophenol and examined using Olympus BX50 Fluorescence Microscope with digital camera (Olympus optical Co., Ltd, Japan), at least 24 conidia were measured for each isolate (Cappuccino and Sherman, 2001).
Appressoria were produced by a slide culture technique (Sutton, 1980). For that, small square of PDA media was cut into 10 × 10 mm2 pieces and settled on vacant Petri dish. A small spore fragment taken from a sporulating fungal culture was distributed on the surface of this small square piece of PDA media and promptly concealed with a cover slip. After 14 days, the cover slip which formed the appressoria across the underside of the cover slip and media was detached and placed on a drop of Lacto phenol on a glass slide. Approximately 20–30 appressoria were microscopically monitored for each isolate.
DNA extraction, PCR amplification, and sequencing
All the selected fungal isolates were grown on V8 juice media for 7 days and genomic DNA was extracted using a protocol as described in Cenis (1992) with minor modifications. The mycelia mat of each isolate were scraped with glass slides and transferred to 1.5 ml Eppendorf tubes. Three hundred microliters of extraction buffer (250 mM NaCl, 200 mM Tris HCl pH 8.5, 0.5% SDS, 25 mM EDTA) was added to fungi filled E-tubes and ground with an autoclavable plastic pestle (8.5 cm). Afterwards, 150 μl of 3 M C2H3NaO2 (Sodium acetate, pH 5.2) was added and tubes were placed at −20°C for about 10 min. Tubes were then centrifuged in a microfuge at 13,000 rpm for 5–10 minutes and the supernatants were transferred to other tubes. Thereafter, an equal volume of isopropanol (isopropyl alcohol) was added and kept for at least 5 min at room temperature. The tubes were placed on ice for 10 minutes and then precipitated DNA was pelleted by centrifugation at 13,000 rpm for 20 minutes. After a wash with 500 μl of 70% ethanol for 3 minutes centrifugation, the pellet was vacuum dried for 5 minutes and dissolved in 50 μl of DW (sterilized distilled water) and stored at −20°C until further use. A sample of each DNA isolate was confirmed by visualizing under ultraviolet illumination with gel electrophoreses and DNA sizes were compared with standard marker.
To confirm the morphological characterization, genomic region were amplified using; an internal transcribed spacer (ITS) rDNA region using universal primers ITS1 and ITS4 (White et al., 1990) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene using primers GDF and GDR (Templeton et al., 1992) respectively. PCR amplification was accomplished using 20 μl reaction volume containing 1 μl of each primer, 1 μl of Genomic DNA (100 ng) and sterilized distilled water. Negative controls containing the same reagents but without DNA template were also involved in all PCR reactions. The amplicons were analyzed by running the samples on a 1% ethidium bromide (EtBr)-stained agarose gels in TAE buffer and the DNA bands were visualized in a UV transilluminator. The PCR products were then purified and directly sequenced with the same primers by a commercial sequencing service provider (Macrogen, Daejeon, South Korea). For all these isolates, sequencing was carried out in both forward and reverse direction to avoid errors. Homology searches for resulting sequences were performed by comparing with those deposited in GenBank Nucleotide database hosted by the NCBI (National Center for Biotechnology Information) using BLAST (Boratyn et al., 2013) and obtaining forward and reverse sequences which were aligned with T-coffee Multiple Sequence Alignment program. All the sequences generated in this study were deposited in GenBank and assigned accession numbers.
Phylogenetic analysis
The phylogenetic analysis was accomplished with multilocus alignment with two combined genes (ITS and GAPDH) using MEGA 7.0 software (Kumar et al., 2016). Sequence similarity searches were performed using the nucleotides BLAST program (http://blast.nih.gov/Blast.cgi) and sequence alignment was manually done with closely related reference sequences of other isolated Colletotrichum species available in NCBI database using ClustalW2 software (http://www.ebi.ac.uk/Tools/phylogenecy/clustalw2-phylogeny/). The reliability of the evolutionary relations in this tree was evaluated with 1,000 bootstrap replications to determine the percentage for each clade. The sequence of C. orbiculare was used as an outgroup in all analyses.
Pathogenicity test
All 18 resultant isolates were tested on two apple cultivars; Tsugaru (Aori) cultivar and Fuji cultivar. For preparation of spore suspensions, the selected isolates were incubated on V8 juice agar media at 25 ± 2°C in the dark for 7 days, as V8 juice media enables more rapid growth and conidia production than PDA media. To obtain the inoculum, the culture was flooded with 10 ml of sterile distilled water and the surface was slightly scraped by microscope glass slide. The suspension was then filtered through two layers of muslin cloth and the conidia concentration was examined and adjusted to 106 conidia/ml using a hemocytometer. For fungal pathogenicity tests, the 18 isolated fungi were inoculated on healthy fruits by wound/drop method at the middle of each fruit and 2 apple cultivars (Aori and Fuji) were used.
For each isolate, 3 healthy fruits were rinsed with tap water, sterilized with 1% sodium hypochlorite (NaClO) solution for 3 minutes and rinsed twice with distilled water. After surface sterilization, the fruits were dried with sterilized tissue paper and placed on a plastic box (28 × 20 × 9 cm; length × width × height) with 3 fruits per box. Then, the apple fruits were pinpricked to 1 mm depth at the middle of each fruit and 10 μl spore suspension (1 × 106 conidia/ml) was pipetted on this wounded spot. All the control fruits were treated with 10 μl of sterilized distilled water. To develop the disease, inoculated fruits were secured in plastic containers lined with wet paper towels and incubated for 14 days at 25°C in constant light. Three fruits were selected per isolate in each cultivar and the experiment was performed twice. Containers were opened once a day to check the development lesions and lesion diameters were measured. Fourteen days after inoculation (DAI), the fruits were cut through the centers of lesions to observe the V-shape symptoms and measure of lesion depth. Isolates were considered pathogenic when the disease lesion area progressed over a 5 mm diameter.
To achieve the Koch’s postulates, the developing symptoms on the inoculated fruits were compared with original symptoms which were observed in the field and the fungi were re-isolated from inoculated fruits following the above method.
Results
Isolates obtained from infected fruits
A total of 18 Colletotrichum isolates were obtained from typical symptomatic fruits acquired from different apple orchards in South Korea (Table 1). From 7 day old colonies of these 18 isolates, Colletotrichum spp. were segregated into 4 morphological subtypes (Type 1, 2, 3 and 4) according to colony characteristics and conidial shape; type 1, 2, 3 and 4, respectively comprised of 3, 2, 8, and 5 isolates. These 4 types shared resemblance to the morphological characteristics of 2 Colletotrichum species that have been frequently correlated with the bitter rot disease of apple, C. acutatum and C. gloeosporioides.
Morphological characteristics
Measurement and appearance of colony characteristics on PDA and V8 juice were described in Table 2. Isolates belonging to Type 1 subtype produced pale yellow colored pigmentation with orange clusters of conidial masses which were submerged in mycelia on PDA. Colonies on V8 juices were smoky grey on the front side and purplish grey on the reverse side. Conidia were 9.5–17 × 4–5.5 μm and fusiform in shape with both ends pointed or slightly acute. Appressoria were clavate to irregular in outline, brown colored, and 6–12 × 4.5–7 μm. The conidial shapes of the isolates in Type 1 resembled those of C. acutatum.

Description of morphological characters of isolated Colletotrichum species based on colony character, conidial shape, and appressoria
Type 2 isolates formed white to grey-colored mycelia with orange-colored conidial masses on PDA media. Colonies on V8 juices appeared white to pale grey-colored on the top and pale orange on the underside. Conidia were 10–18 × 4–6 μm, mostly fusiform in shape, but some were obclavate (both ends pointed or one acute ± rounded). Appressoria were single, medium brown, clavate or irregular in outline, and 5–13 × 3.5–7.5 μm. Type 2 conidia shape resembled those of C. acutatum.
Type 3 isolates formed olive grey to iron grey mycelia in the center of PDA plates. Colonies on V8 juices had light grey mycelium on front view and pale orange aerial mycelium on back view. Conidia were mostly cylindrical in shape with rounded ends, up to 10–15.5 μm × 4–6.2 μm. Appressoria were ovoid to slightly irregular in shape, dark brown in color, and ranged from 6–12 μm × 4–7 μm. The features of the isolates in Type 3 were similar to those of C. gloeosporioides.
Isolates of Type 4 formed cottony grey-colored colonies with abundant mycelia on PDA media. Colonies on V8 juices had a white to milky-fluffy mycelium on top view and the underside was pale yellow. Conidia in all isolates were 10.5–16 × 3.5–5 μm and had cylindrical-shape with slightly rounded (sometimes oblong) ends. Appressoria were clavate, brown to dark brown in color, ovoid and slightly irregular in shape, with dimension of 5.5–11 × 3.5–7.5 μm. Based on these morphological features, the isolates in Types 4 resembled the description of C. gloeosporioides. The description of morphological characteristics for the Colletotrichum species in Type 1–4 is shown in (Fig. 1A, 1B).
Molecular characteristics and Phylogenetic analysis
The ITS and GAPDH regions of all isolated fungi were amplified and sequenced. The PCR products obtained from ITS and GAPDH gene sequence of isolated fungi were approximately 550 and 250 base pairs (bp) long respectively and homology searches with each individual sequence suggested that all tested isolates corresponded to the genus Colletotrichum. The sequences of all isolates within the same type were 100% identical to each other with no evident differences.
Based on the sequence comparison of all the obtained isolates with published sequences in GenBank, isolates in Type 1 matched well with sequences belongings to C. fioriniae, the isolates in Type 2 shared homology to C. nymphaeae, all isolates in Type 3 had sequence identity to C. siamense and the sequences from isolates in Type 4 corresponded to the sequences from C. fructicola.
Molecular phylogenetic analyses based on final sequence alignment of a combined data set of ITS and GAPDH regions in a separate analysis classified the Colletotrichum isolates into two major clades, corresponding to the C. acutatum species complex and C. gloeosporioides species complex, with bootstrap support of 100% (Fig. 4). The C. acutatum species complex clade included two separate subclades: C. fioriniae and C. nymphaeae. The C. gloeosporioides species complex clade comprised of two sub-clades: of C. siamense and C. fructicola. Notably, this is the first time that these 4 species have been identified as the causal agents of bitter rot disease on apple in South Korea.
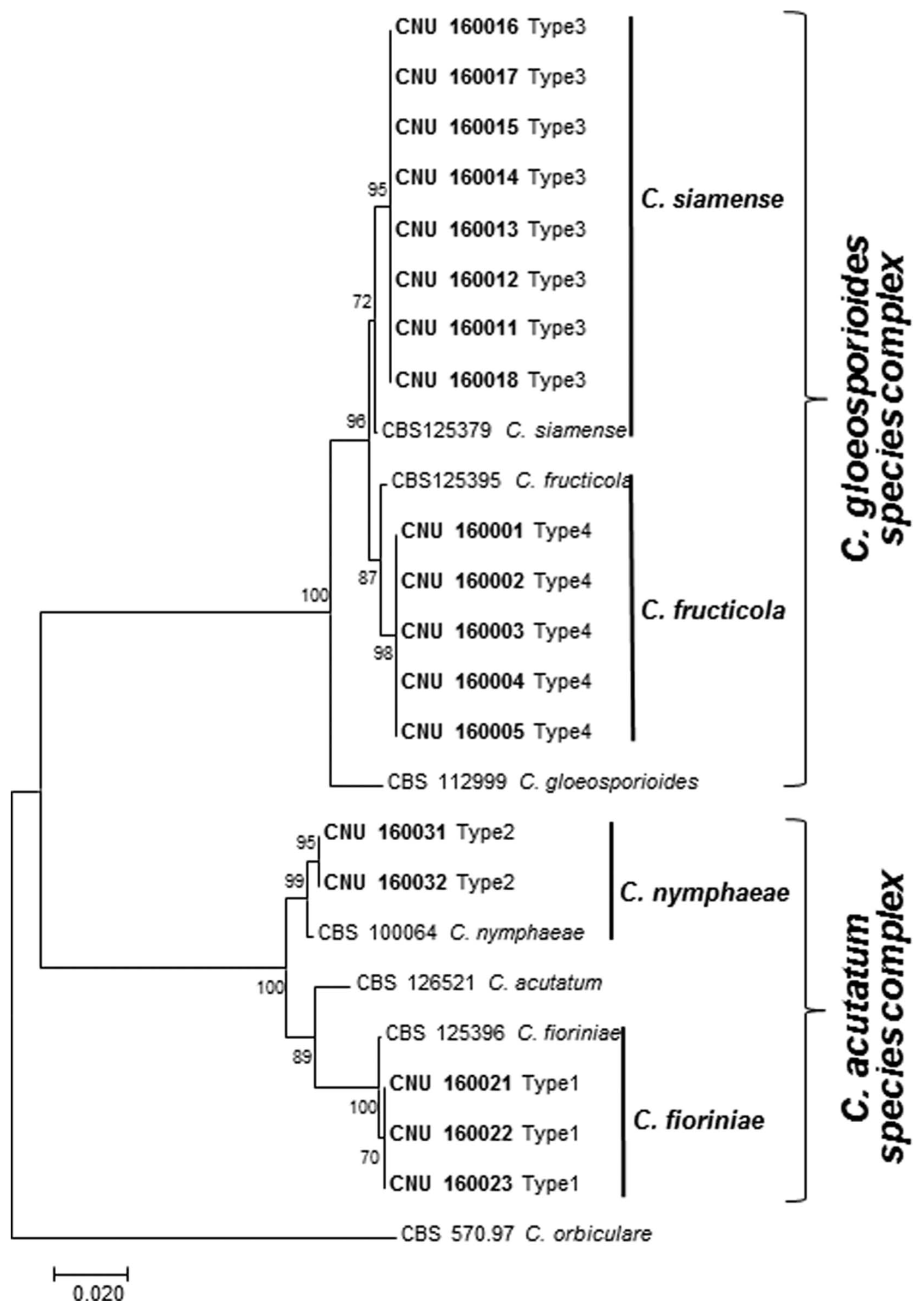
Maximum parsimony analysis showing phylogenetic relationships among 18 isolates of Colletotrichum from apple and other related Colletotrichum spp. based on combined dataset of internal transcribed spacer (ITS) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene sequences. Numbers beside each branch represent bootstrap values obtained after a bootstrap test with 1,000 replications. Present isolates are shown in boldface. Bar indicates the number of nucleotide substitutions. The tree is rooted with Colletotrichum orbiculare.
The representative ITS and GAPDH genetic sequences obtained from the isolates were deposited in GenBank with accession numbers given in Table 1. The final sequence alignment of combined datasets of ITS and GAPDH containing 25 taxa (including current 18 isolates and 7 reference sequences from GenBank were used to compare the tree output. All Colletotrichum isolates resulted from the current study. The species used in this study are presented in Table 1.
Pathogenicity test
All 18 isolates of Colletotrichum species were pathogenic and produced the typical bitter rot symptoms on inoculated-apple fruits belonging to Fuji and Aori cultivars (Fig. 2A). These symptoms consisted of sunken lesions with orange to pink-colored acervuli arranged concentrically. V-shaped necrotic tissue zones were also observed when the fruits were cut through the centers of lesions (Fig. 3). The Fuji cultivar developed the lesion symptom earlier than that of Aori cultivar. All the isolates belonging to the C. gloeosporioides species complex, formed bigger and deeper lesions than the isolates belonging to the C. acutatum species complex isolates (Fig. 2B). Fruit lesion diameters of the 18 isolates are described in Table 3. Among all 18 isolates, Type 3 isolates (C. siamense) showed the widest and deepest lesions (43.5 mm in diameter and 30.5 mm in depth on Fuji cultivar; 39.5 mm in diameter and 25.5 mm in depth on Aori cultivar) two week after inoculation (Fig. 2). After C. siamense, Type 4 isolates (C. fructicola) produced the second largest lesion among these 4 species (35.5 mm in diameter and 25.5 mm in depth on Fuji cultivar; 32.5 mm in diameter and 25.0 mm in depth on Aori cultivar). In the C. acutatum species complex (C. nymphaeae and C. fioriniae), the lesion produced by the Type 1 isolates (C. fioriniae) was a slightly larger (25.5 mm in diameter and 17.5 mm in depth on Fuji cultivar; 18.5 mm in diameter and 15.5 mm in depth on Aori cultivar) than that produced by Type 2 isolates (C. nymphaeae) (20.5 mm in diameter and 16.5 mm in depth on Fuji cultivar; 16.5 mm in diameter and 14.0 in depth on Aori cultivar). However, the depth of the lesion caused by these two species was not significantly different. No symptoms were observed on control fruits treated with sterile distilled water (Fig. 2A). Consistent results were obtained in the replicated experiments.
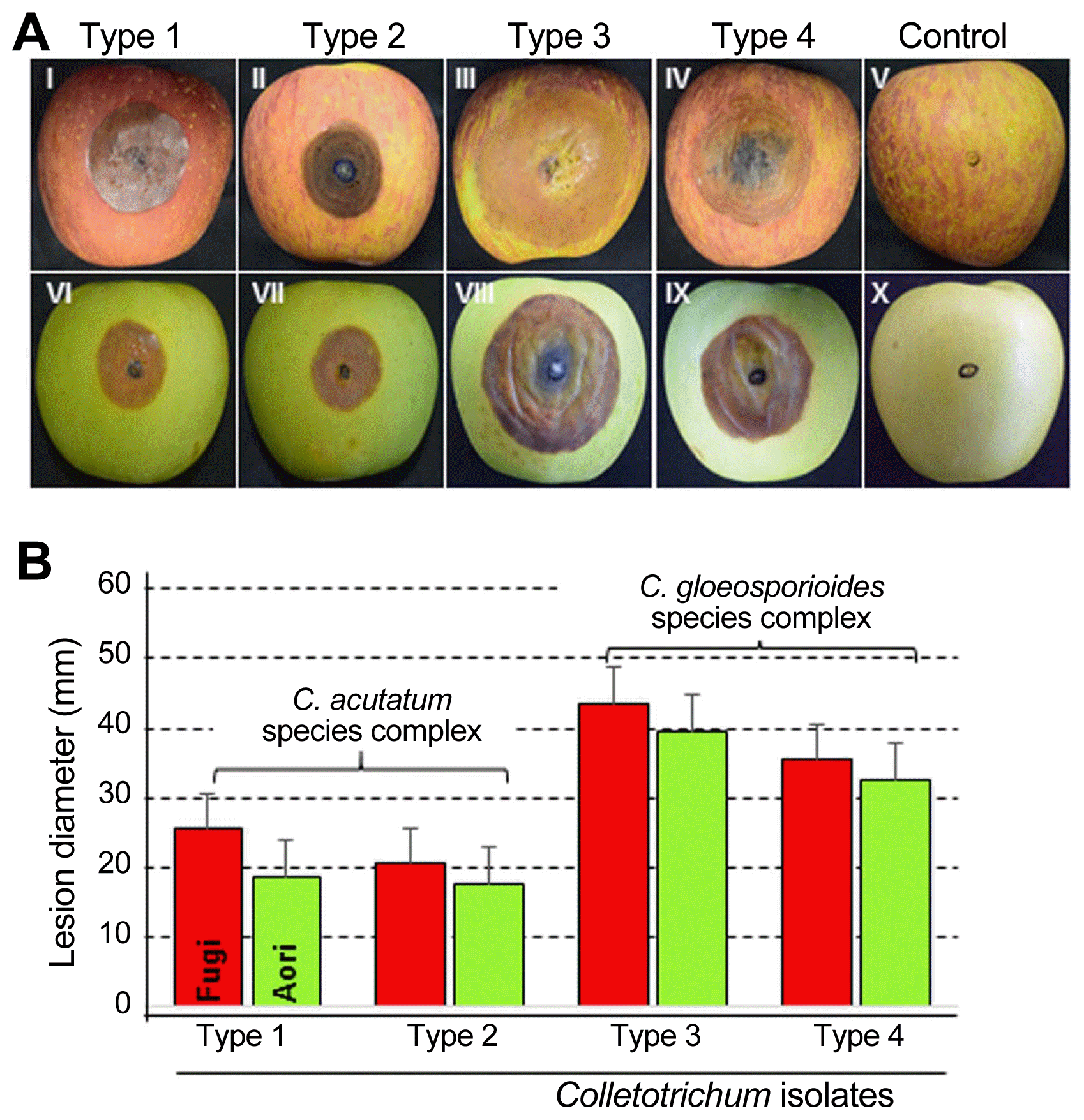
(A) Symptoms on apple fruits after wound/drop inoculation with Colletotrichum spp. Symptoms produced by (I, IV) Type 1, C. fioriniae; (II, VII) Type 2, C. nymphaeae; (III, VIII) Type 3, C. siamense; (IV, IX) Type 4, C. fructicola; (V, X) control fruits. Fruits (I–V) on Fuji cultivar and (VI–X) on Aori cultivar. Lesions were 14 days after inoculation (DAI). (B) Comparison of lesion length among four types of Colletotrichum species on Fuji and Aori cultivars.
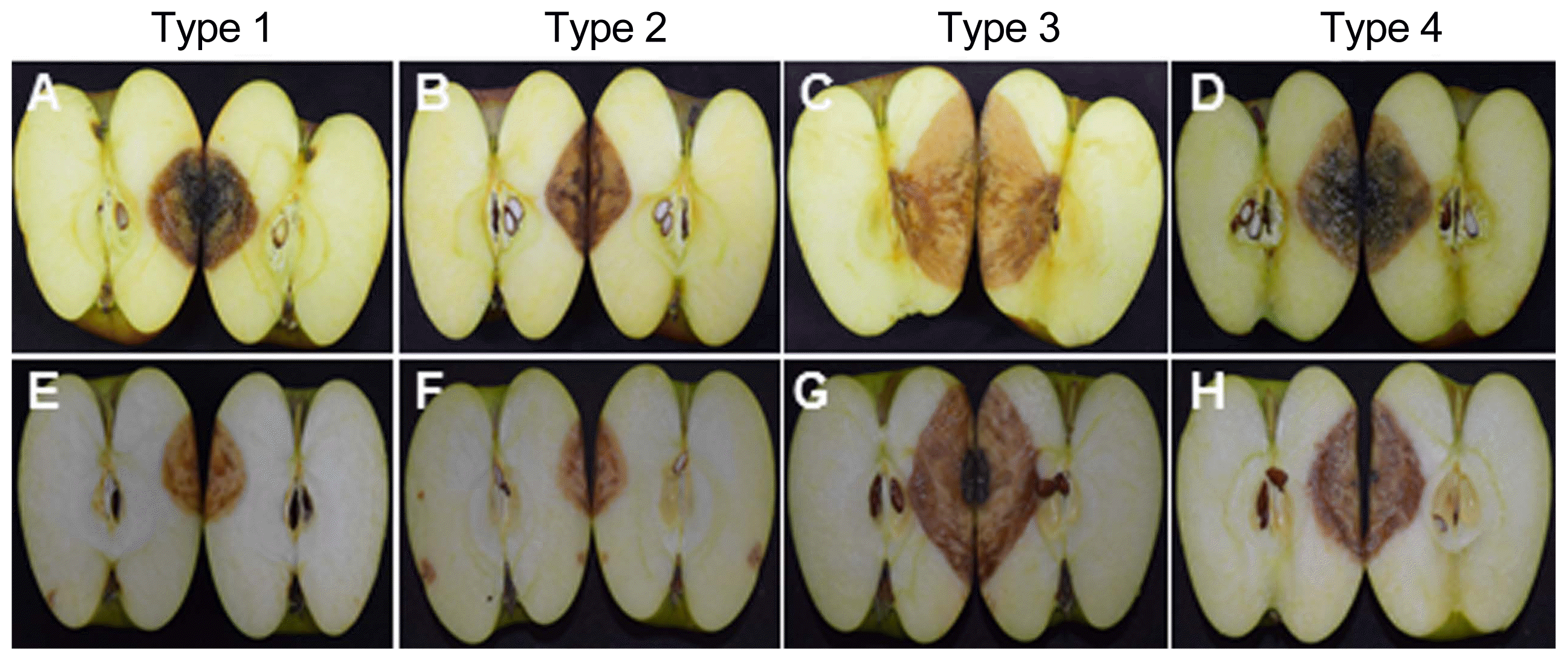
V-shaped necrotic tissues by cause of wound/drop inoculation of (A, E) Type 1, C. fioriniae; (B, F) Type 2, C. nymphaeae; (C, G) Type 3, C. siamense; (D, H) Type 4, C. fructicola on (A, B, C, D) Fuji cultivar and (E, F, G, H) Aori cultivar. Necrotic tissues symptoms were 14 days after inoculation (DAI).
Discussion
The current study is focused on the Colletotrichum species isolates obtained from apple fruits afflicted with bitter rot disease in South Korea. Based on the morphological and molecular characteristics (including ITS and GAPDH gene sequences) the resulting 18 isolates were divided in 4 groups: Type 1 (3 isolates), Type 2 (2 isolates), Type 3 (8 isolates) and Type 4 (5 isolates) as described in Table 1. Thus, the present study showed that the isolates of Colletotrichum spp. causing the bitter rot disease in Korea belong to 4 species: C. fioriniae (Type 1); C. nymphaeae (Type 2); C. siamense (Type 3); C. fructicola (Type 4) where C. fioriniae and C. nymphaeae belong to Clade 3 and Clade 2 respectively of C. acutatum species complex and C. siamense and C. fructicola belong to Musae Clade of C. gloeosporioides complex species. Interestingly, to the best of our knowledge, this is the first report that implicates these 4 species in the pathogenesis of bitter rot disease in South Korea. Therefore, the population Colletotrichum species associated with Apple bitter rot disease in South Korea maybe more complex than was previously recognized.
According to the morphological result, our result also showed that the isolates of C. gloeosporioides complex species grew faster than that of C. acutatum complex species isolates on PDA and V8 juices media. Generally, spore shape has traditionally been used for differentiating C. acutatum from C. gloeosporioides. However, identifying species within the Colletotrichum genus using morphological characteristics is challenging because of increasing complexity and morphological similarity within different members of Colletotrichum species (Park et al., 2018). The morphological characteristics are ambiguous, and indefinite morphotaxonomic characters tend to vary depending on the experimental method used and conditions at the time of experiment (Oo et al., 2016). Thus, the novelty of this study is the combination of morphological characteristics and molecular traits to distinguish different species of Colletotrichum and confirm the identity of extracted isolates. The present study showed that morphological characteristics alone could not distinguish the species within a species complex as these features could discriminate only the C. acutatum species from C. gloeosporioides species. Identification of Colletotrichum species using species-specific primers was found to be useful as it gave reliable and replicable results. Even though the ITS sequences have been widely used in identification of Colletotrichum species, they could not clearly differentiate the species within species complex. GAPDH sequences, on the contrary, gave the accurate differentiation of Colletotrichum species within C. acutatum and C. gloeosporioides complex species. The multi-sequence alignment using ITS sequence data did not show the conflict in tree topology suggesting that the tree sequence datasets of ITS and GAPDH could be combined for further analysis. Therefore, the results derived from combined molecular approaches provided more reliable identification method to distinguish Colletotrichum species.
Pathogenicity tests showed that all the tested isolates from four Colletotrichum species were pathogenic to wounded apple fruits and lesion sizes did not differ in the two tested apple cultivars, Aori and Fuji cultivars (Fig. 2A). However, pathogenic comparison between these 4 species showed that the current isolates belonging to the C. gloeosporioides species complex (Type 3 and Type 4 isolates; C. siamense and C. fructicola) produced larger lesions than isolates belonging to the C. acutatum species complex (Type 1 and Type 2 isolates; C. fioriniae and C. nymphaeae). Additionally, C. siamense in the C. gloeosporioides complex species produced the biggest, widest lesions and was found to be the most aggressive among these 4 species. Even though the current 18 isolates are not enough to represent all the species causing bitter rot disease in Korea, there is a high probability that isolates (C. siamense and C. fructicola) belonging to C. gloeosporioides complex species is the dominant pathogen in Korea as compared to the other isolates (C. fioriniae and C. nymphaeae) in C. acutatum complex species (Fig. 4). Therefore, the current result is consistent with a previous report (Munir et al., 2016).
In conclusion, our results indicate that accurate identification of pathogens within a species complex of genus Colletotrichum is required for management of bitter rot disease on apple fruit. Moreover, these results emphasized molecular phylogenetic analysis and pathogenicity tests along with the morphological identification are better tools for the correct identification and characterization of the Colletotrichum species.
Acknowledgments
The authors would like to thank Prof. Jun Myoung Yu (Chungnam National University) for critically reviewing the manuscript. This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (Project No. NRF-2014R1A1A2057824), Republic of Korea.