Visualization of Phytophthora palmivora Infection in Oil Palm Leaflets with Fluorescent Proteins and Cell Viability Markers
Article information
Abstract
Bud rot (BR) is the most devastating disease affecting oil palm (Elaeis guineensis) crops in Colombia. Its causal agent, Phytophthora palmivora, initiates the infection in immature oil palm leaflets producing necrotic lesions, followed by colonization of opportunistic necrotrophs, which increases disease damage. To improve the characterization of the disease, we transformed P. palmivora using Agrobacterium tumefaciens-mediated transformation (ATMT) to include the fluorescent proteins CFP-SKL (peroxisomal localization), eGFP and mRFP1 (cytoplasmic localization). The stability of some transformants was confirmed by Southern blot analysis and single zoospore cultures; additionally, virulence and in vitro growth were compared to the wild-type isolate to select transformants with the greatest resemblance to the WT isolate. GFP-tagged P. palmivora was useful to identify all of the infective structures that are commonly formed by hemibiotrophic oomycetes, including apoplastic colonization and haustorium formation. Finally, we detected cell death responses associated with immature oil palm tissues that showed reduced susceptibility to P. palmivora infection, indicating that these tissues could exhibit age-related resistance. The aim of this research is to improve the characterization of the initial disease stages and generate cell biology tools that may be useful for developing methodologies for early identification of oil palm materials resistant or susceptible to BR.
Introduction
Palm oil represented approximately 34% of the total vegetable oil produced worldwide in 2015, exceeding the oil produced from other crops, such as soybean, sunflower and rapeseed (Corley and Tinker, 2015). The main producers worldwide are Malaysia and Indonesia, and Colombia was the first producer in the Americas with 466185 ha of oil palm that produced 1272523 ton of crude oil in 2015 (Corley and Tinker, 2015; Fedepalma, 2015). The main disease that affects oil palm (Elaeis guineensis Jacq.) crops in America is the bud rot disease (BR) caused by the oomycete Phytophthora palmivora Butl. (Drenth et al., 2013; Martínez et al., 2010; Sarria et al., 2008; Torres et al., 2010). Up to 2014, this disease caused the eradication of more than 70000 ha of crops in Colombia, and the economic losses escalated to approximately 250 million USD (reviewed by Torres et al., 2016).
Since 2008, when P. palmivora was identified as causal agent of BR, research examining this topic has been focused on the development of methods to discover oil palm-resistant materials suitable for plant breeding, and on identifying early symptoms and biochemical responses to improve agronomical practices to control the disease (Martínez, 2009; Moreno-Chacón et al., 2013; Navia et al., 2014). Sarria et al. (2015) produced the first histopathological description of the disease using optical and scanning electron microscopy in detached leaflets, with a precise description of the penetration process, appressorium formation and further emergence and sporulation in the necrotrophic phase; however, in vivo studies, apoplast colonization and haustorium formation are difficult to observe using these methodologies.
To improve the histological characterization of oomycetes during the colonization of plant tissues and to provide more detailed analyses using cell and molecular biology techniques, different methodologies of genetic transformation have been developed combined with the utilization of green fluorescent protein (GFP) and its derivatives (Judelson and Ah-Fong, 2009). Genetic transformation of different Phytophthora species has been achieved using protoplast transformation (Bottin et al., 1999; Judelson et al., 1991, 1993; Si-Ammour et al., 2003), electroporation (Huitema et al., 2011; Latijnhouwers et al., 2004), particle bombardment (Cvitanich and Judelson, 2003) and Agrobacterium tumefaciens (Vijn and Govers, 2003; Wu et al., 2016a); however, selection of the most appropriate technique is difficult because the success of each protocol may vary intra and interspecifically (Judelson and Ah-Fong, 2009). Hence, different transformation protocols must be tested for the genetic transformation of each Phytophthora isolate.
Genetic transformation of Phytophthora spp. was developed for diverse applications, including the quantification of resistance levels in potato cultivars (Kamoun et al., 1998), evaluation of chemical inducers of disease resistance (Si-Ammour et al., 2003), gene silencing for functional characterization (Blanco and Judelson, 2005; Bos et al., 2010; van West et al., 1999a), cell biology studies of pathogen structures (Ah-Fong and Judelson, 2011; Bozkurt et al., 2011; Chaparro-Garcia et al., 2011; Dunn et al., 2013; Schornack et al., 2010) and more recently, genome editing using the CRISPR/cas9 system (Fang and Tyler, 2016).
Transformation in P. palmivora has been accomplished using three of four of the abovementioned methodologies. Protoplast transformation in the P6390 isolate with GFP and GUS markers (van West et al., 1999b), A. tumefaciens-mediated transformation in the P6390 isolate with the gene encoding for the enzyme β-glucoronidase and the and the P1 isolate with the (Vijn and Govers, 2003; Wu et al., 2016a) and zoospore electroporation in the P6390 and P16830 isolates with TD-tomato and YFP-KDEL (tagging the endoplasmic reticulum), respectively (Rey et al., 2015). These fluorescently tagged isolates have been used to detect different Phytophthora palmivora structures in Medicago truncatula, and Hordeum vulgare (Le Fevre et al., 2016; Rey et al., 2015).
The aim of this research was to select the best fluorescent protein for tagging P. palmivora isolates obtained from oil palms affected by BR, in order to provide a detailed characterization of both the biotrophic and necrotrophic stages of the infectious process. We successfully transformed three isolates of P. palmivora by A. tumefaciens-mediated transformation, but the most amenable isolate to this procedure was CPPHZC-05. The transformants tagged with the fluorescent proteins allowed us to detect all of the common structures during the biotrophic and necrotrophic stages as well as the penetration of zoospores in immature oil palm leaflets. Moreover, the use of cell viability markers allowed us to visualize cell death patterns during the infectious process that were associated with age related plant resistance.
Materials and Methods
Phytophthora palmivora isolates
The Colombian isolates of P. palmivora, CPPhZC-05, CPPhZOC-03 and CPPhZN- 02 collected from oil palm affected tissues, were acquired from the CENIPALMA microorganism collection (research authorization number 269 of 2013 given by the Autoridad Nacional de Licencias Ambientales ANLA). All isolates were maintained in solid clarified 20%-V8 agar (Erwin and Ribeiro, 1996) supplemented with 100 μg/ml carbenicillin.
Plasmid construction
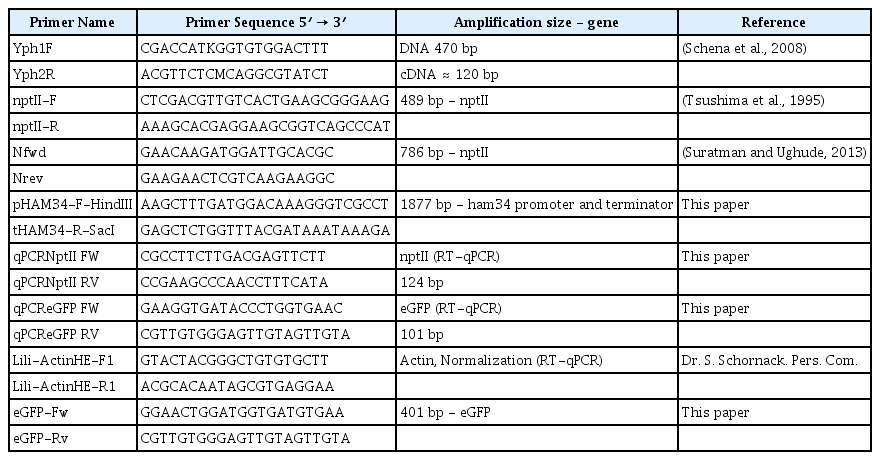
The binary plasmid pNPTII was donated by Dr. Francine Govers from Wageningen University (Netherlands) (Vijn and Govers, 2003). To include different fluorescent protein genes flanked by the Ham34 promoter and terminator in the pNPTII vector, we designed a forward primer in the 5′ region of the Ham34 promoter containing the HindIII restriction site and a reverse primer in the 3′ region of the Ham34 terminator containing the SacI restriction site (Table 1). The templates for the PCR reaction were the vectors pGFPH, pCFP-SKL and pTOR-mRFP1, which contained fluorescent protein genes flanked by the Ham34 promoter and terminator. The pGFPH and pCFP-SKL plasmids were kindly provided by Dr. Howard Judelson of UC Riverside, USA (Ah-Fong and Judelson, 2011), http://oomyceteworld.net/TransformationMain.html). The obtained amplicon, was sub-cloned in PCR®4 TOPO® (Invitrogen, Carlsbad, CA, United States), and then the fluorescent protein-Ham34 fragment was excised with HindIII and SacI fast digest restriction enzymes (Thermo Scientific™, Waltham, MA, USA) and ligated into the pNPTII vector using a T4 DNA ligase (Thermo Scientific ™, Waltham, MA, USA). All plasmids were confirmed by ABI sequencing at Macrogen, Inc. (Seoul, South Korea). The general structure of the T-DNA is shown in Fig. 1A.
(A) Schematic representation of the T-DNA included in the binary plasmid pNPTII. FP: Fluorescent protein. (B) Representative evaluation of transformant colonies by Southern blot with nptII probe three months after transformation; a wild type (WT) isolate was used as a negative control. Dashed lines show the colonies selected for monosporic cultures. The bottom blot shows the hybridization obtained from colonies derived from different single zoospore cultures.
Agrobacterium tumefaciens strains and culture conditions
The genes coding for the fluorescent proteins cloned in the pNPTII vector were electroporated into A. tumefaciens strains AGL1, GV3101 and LBA4404. Five days before agroinfection, we inoculated 2 plates of minimal medium (Hooykaas et al., 1979) plus 50 μg/ml kanamycin and appropriate antibiotics for each A. tumefaciens strain. The cultures were incubated at 25°C in the dark for four days. One day before agroinfection, 10 ml of liquid minimal medium supplemented with the same antibiotics as the plates, were inoculated with some of the colonies obtained in the petri dish and incubated overnight at 25°C and 300 rpm in the dark. On the day of agroinfection, the culture was centrifuged in a swinging bucket rotor centrifuge at 5000 g, and the bacterial pellet was washed twice with induction medium (minimal medium supplemented with 10 mg/l β-sitosterol, 7.8 g/l MES, 0.1% glycerol and 200 μM acetosyringone; pH 5.2–5.3). Finally the bacterial pellet was resuspended in 10 ml of induction medium and incubated for 5 h at 25°C and 300 rpm in the dark (adapted from Vijn and Govers, 2003).
Phytophthora palmivora transformation (adapted from Vijn and Govers, 2003)
Seven P. palmivora culture plates in 15-mm petri dishes were flooded with 10 ml of ice-cold 1 X petri’s solution, and the mycelia was exhaustively rubbed with a Drigalski spatula to harvest the sporangia (Huitema et al., 2011). The solution was recovered and incubated in a tridimensional shaker under constant light for approximately 30 min until most of the zoospores were liberated. The zoospore concentration was determined using a Neubauer chamber and adjusted to a final volume of 30 ml at a concentration of approximately 106 zoospores/ml. The zoospore suspension was inoculated with 2 ml of an A. tumefaciens culture as described above at an OD600nm of 0.45. The zoospore suspension was incubated at room temperature for 30 min, and then it was vigorously shaken to induce zoospore encystment and then incubated under the same conditions for another 90 min. The cysts were centrifuged for 5 min at 1500 g in a swinging bucket rotor centrifuge and resuspended in a final volume of 5 ml. One milliliter of the suspension was spread over induction medium plates covered with Amershan HybondTM N+ membranes (GE Healthcare, UK). The plates were incubated at 25°C in the dark for 5 days. The nylon membranes were cut into 16 pieces and placed upside down over Plich medium selection plates plus 200 μg/ml cefotaxime and 7.5 μg/ml G418 for the CPPhZC-05 and CPPhZN-02 isolates or 12 μg/ml for the CPPhZOC-03 isolate. After 7 days, transformant colonies were transferred to solid clarified 20%-V8 agar plus 200 μg/ml cefotaxime, 25 μg/ml vancomycin and 16 μg/ml G418 for CPPhZC-05 and CPPhZN-02 isolates or 20 μg/ml for the CPPhZOC-03 isolate.
Molecular evaluation of transformant colonies
Transformant colonies were grown in a 250-ml Erlenmeyer containing 100 ml of liquid 20%-V8 clarified medium plus 200 μg/ml cefotaxime, 25 μg/ml vancomycin and 16 μg/ml G418 for the CPPhZC-05 and CPPhZN-02 isolates or 20 μg/ml for the CPPhZOC-03 isolate. The cultures were incubated for 1 week at 28°C and 150 rpm. The biomass produced was filtrated in sterile gauze and pulverized in liquid nitrogen with a mortar and a pestle. This biomass was used for DNA and RNA extraction.
The DNA for PCR was extracted using the GeneJET plant genomic DNA purification kit (Thermo scientific, Waltham, MA, USA). The PCR reactions were elaborated in a final volume of 10 μl containing a concentration of 1 X PCR buffer, 2 mM MgCl2, 1 mM dNTPs, 0.5 μM primers, 0.05 u/μl Taq polymerase (Thermo scientific, Waltham, MA, USA) and 20 ng of template DNA. The primers used for all procedures are listed in Table 1. All transformant colonies were evaluated for nptII and the presence of GFP with Ypt1 as a control for amplification.
Southern blotting
DNA extraction for Southern blotting was performed according to Goodwin et al. (1992) with an additional purification step with CTAB buffer and phenol:chlorophorm:isoamyl 25:24:1. The genomic DNA concentration was estimated using a Biotek synergy MX instrument (Winooski, VT, USA). Twenty-five micrograms of DNA were digested with EcoRI fast digest restriction enzyme for 18 h (Thermo Scientific™), and the digestion products were separated in a 0.9% agarose gel with 1 X TAE buffer. The DNA was transferred to a positively charged nylon membrane (Roche diagnostics, Mannheim, Germany) by capillary transfer in 20 X SSC buffer. DIG-labelled nptII probe was synthetized using primers Nfwd-Nrev (Table 1) and hybridized at 42°C for 16 h. Chemiluminescence was performed with CSPD and chemiluminescent detection films according to manufacturer’s instructions (Roche diagnostics, Mannheim, Germany).
qRT-PCR of GFP in transformant colonies
The RNA extractions were performed using the RNeasy Plant Mini Kit (Qiagen®, Hilden, Germany). One microgram of RNA was treated with amplification grade DNAse I (Invitrogen, Carlsbad, CA, United States); the amount of DNase was increased to 1.5 μl in comparison to the manufacturer’s instructions. Prior to cDNA synthesis, PCR of the Ypt1 gene using RNA as a template was performed to confirm that the DNAse treatment was successful.
The first-strand cDNA was synthetized with Super- Script™ IV Reverse Transcriptase (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The quality of the cDNA was first evaluated with primers Yph1F-2R, which amplify fragments of 470 bp and ≈120 bp using DNA and cDNA as template, respectively.
Real-time PCR reactions for relative quantification were performed in a final volume of 15 μl containing 1 X Evagreen master mix (Biotium, Hayward, CA, USA), 0.67 μM of each primer and 5.5 μl of a 1:5 cDNA dilution. The reactions were assessed in a LightCycler® 480 real time PCR instrument (Roche diagnostics, Mannheim, Germany). Estimation of the expression ratio was calculated using the basic quantification mode of the LightCycler® 480 software, release 1.5.0.
Monosporic cultures
Two different transformant colonies (025 with three integrations and 036 with two integrations) were grown in selective V8 clarified agar for 10 days in 15-mm petri dishes. Each plate was flooded with 15 ml of ice-cold 1 X modified petri’s solution without rubbing the mycelia. Zoospore liberation was monitored for approximately 40 minutes, and then 1 ml of the zoospore suspension was collected to make 1:10 serial dilutions in 1x modified petri’s solution. One hundred microliters of each dilution were plated in selective V8 clarified agar and verified under the stereomicroscope to choose the single zoospores. Colonies were transferred to fresh clarified V8 selective plates. All monosporic cultures were validated by qRT-PCR and Southern blotting as previously described.
Leaflet inoculation and fluorescence microscopy
Immature leaflets were inoculated as previously reported by Velez et al. (2013) and Sarria et al. (2015). Briefly, immature leaflets from a 13-year-old oil palm Golden Hope cultivar were dissected into 10-cm-long pieces and introduced to humidity chambers. The leaflets were inoculated with a 20-μl droplet of a suspension of 60,000 zoospores/ml, of the transformant 036. Microscopic observations were performed each hour during the first six hours and then at 12, 18, 24 and 48 h. The inoculated tissue was mounted on glass slides with cover slips and a 60-μl drop of 0.5 μg/μl p-phenylene diamine. The observations were performed using a LEICA DMI4000B inverted fluorescence microscope using bright field and the GFP filters (excitation filter: BP470 nm/40 nm, dichromatic mirror: 500 nm and barrier filter: 525/50 nm). Images were obtained with a LEICA DFC310FX camera, independently for each filter and merged using ImageJ version 1.45S with adjustments for brightness and contrast. Visualization of the cell viability markers was conducted with propidium iodide (PI) and fluorescein diacetate (FDA) as described by Jones et al. (2016).
The GFP filter described above was used to observe the GFP transformants and oil palm cells stained with FDA, the TXRD filter (excitation filter: BP560 nm/40 nm, dichromatic mirror: 595 nm and barrier filter: 645/75 nm) for the mRFP1 transformants and the PI fluorescence observations, and the CFP/YFP filter (excitation filter: BP436 nm/12 nm, dichromatic mirror: 445 nm and barrier filter: 467/37 nm) for the CFP-SKL transformants.
Colony growth assay
Young mycelial plugs of five P. palmivora transformants and the wild type isolate were placed in the center of a 15-mm-diameter petri dish containing V8-clarified agar supplemented with 200 μg/ml cefotaxime and 25 μg/ml vancomycin. The plates were incubated at 28°C in the dark for seven days. The mycelia were scanned, and the colony area was estimated using Photoshop PS6. The colony diameter was estimated with D = (4A/π)1/2, modeling the colony as a perfect circle, where A is the colony area and D is the colony diameter. For each transformant, we generated ten technical replicates in a completely randomized design. Comparisons of the transformants with the WT isolate were performed using a Dunnett’s test with S.A.S. software version 9.1.
Lesion length assay
Inoculated leaflets were prepared as mentioned above. A total of six leaflet pieces of 10 cm were placed in each chamber, and therefore every chamber contained a mock, a wild type isolate and four different transformants. The inoculated leaflets were maintained for four days at 25°C. Each leaflet was scanned, and the lesion length was measured using ImageJ 1.50i. For each transformant, eight replicates were assessed in a randomized complete block design, in which each humidity chamber represented a block. Comparisons of the transformants with the WT isolate were conducted using a Dunnett’s test with S.A.S. software version 9.1.
Results
Plasmid constructs and transformation of P. palmivora with fluorescent protein genes
We were unable to obtain stable transformants in our laboratory using zoospore electroporation, despite performing multiple experiments using three different P. palmivora isolates, with varying electroporation conditions or selection markers (data not shown). The alternative tested methodology was the previously developed Agrobacterium tumefaciens-mediated transformation (ATMT) (Vijn and Govers, 2003). Because the binary plasmid pNPTII does not contain genes encoding fluorescent proteins, we cloned the eGFP, mRFP1 and CFP-SKL genes flanked by the ham34 promoter and terminator of the oomycete Bremia lactucae (Fig. 1A).
For the transformation experiments, we tested three A. tumefaciens strains, AGL1, LBA4404 and GV3101, but it was only possible to obtain colonies that showed normal growth on selective plates with AGL1. Moreover, we obtained transformed colonies with empty vector as well as those containing fluorescent protein genes (Table 2). Among the 89 transformed colonies, 83 were from the isolate CPPhZC-05, but it was possible to obtain a few transformed colonies of CPPhZOC-03 and CPPhZN-02, indicating that this protocol has the potential to be adapted to multiple isolates.

Number of transformant colonies obtained, discriminated by isolate, fluorescent protein and copy number determined by Southern blot
All of the obtained transformed colonies were confirmed by PCR of the selection marker and the respective fluorescent protein in each case. Only one of the transformants carrying the plasmid containing the gene coding for CFP-SKL protein amplified the selection marker gene but not the fluorescent protein gene, indicating that this might be a truncated version of the T-DNA (data not shown). The other transformants showed complete integration of the T-DNA, grew on selection medium and expressed both the selection marker and fluorescence protein as shown by qRT-PCR analyses.
Concerning the stability of transformation, approximately 40% of the transformed colonies lost their viability through successive passes every two weeks on selective plates for 12 months, even though the Southern blot results indicated that the T-DNA was integrated in the P. palmivora genome. The Southern blot analysis showed that nearly all of the evaluated colonies obtained after a single transformation had different band patterns and single or multiple T-DNA insertions, indicating that we obtained different integration events. In addition, most of the transformants had one or two T-DNA insertions. Taken together, these results showed that even after verifying the integration of T-DNA into the genome, there may be a loss of viability in the transformed colonies; however, the cause of this phenomenon remains to be elucidated.
Young mycelia from five-day-old culture plates were observed under a fluorescence microscope to assess both the intensity and the distribution of fluorescence in each transformant. For transformants carrying the proteins mRFP1 and eGFP, the fluorescence was evenly distributed throughout the mycelium, sporangia and chlamydospores because these proteins have a cytoplasmic localization; however, the mRFP1-tagged were brighter than the eGFP-tagged zoospores. Some transformants showed an uneven distribution of fluorescence or no fluorescence at all, but the expression level measured by qRT-PCR could not be clearly correlated with the observed fluorescence (data not shown). Transformant colonies tagged with the CFP-SKL protein, which has a peroxisomal targeting motif, displayed a fluorescence pattern consisting of small dots distributed along the hyphae, sporangia and chlamydospores (Fig. 2). Because the A1 mating type has not been found in Colombia, it was not possible to verify the fluorescence of the oospores (Torres et al., 2010).
Monosporic cultures and heritability
To verify the transfer of T-DNA to the zoospores, we elaborated five single zoospore cultures from the transformed colonies 025 and 036 containing the eGFP gene. Southern blot analysis of the monosporic cultures revealed the same band pattern observed in the initial transformed colony, indicating that the T-DNA is inheritable from the initial transformed cultures (Fig. 1B). Additionally, the expression of eGFP gene was measured by qRT-PCR, but some variations were observed in the expression levels of the tested colonies, indicating changes in the expression of the genes encoding for fluorescent proteins in colonies from different zoospores (Supplementary Fig. 1).
Growth and infectivity of transformants
To verify that the transformed isolates had the same characteristics as the wild type CPPhZC-05, we compared their growth in culture medium as well as their infectivity in oil palm leaflets. By measuring the diameter of the colony, it was determined that three out of five of the tested transformants had the same growth rates in culture as the untransformed control, and only transformant 010 presented a reduced growth rate compared with the non-transformed control. Consequently, the appearance of colonies of this transformant exhibited abnormalities compared with wild type. Transformant 036 showed a slightly increased growth rate compared with wild type (Table 3).
To assess whether the virulence of the transformants was affected by the transformation process, we inoculated oil palm leaflets with transformants carrying GFP (032 and 036), mRFP1 (079) and CFP-SKL (021) derived from the CPPhZC-05 genetic background. The length of the lesion observed between different transformants was not significantly different compared with the wild type CPPhZC-05 (Table 3). Taken together, these results indicate that most of the evaluated transformant colonies were not affected by the transformation process in terms of virulence or the ability of the colony to grow in vitro.
Histological visualization during infection of immature oil palm leaflets
Immature oil palm leaflets that were closer to the meristematic zone were dissected to perform the visualization experiments because they are thin and thus allowed better observation under the fluorescence microscope. During the first 2 h of monitoring, we observed cyst germination and appressoria on the leaflet surface as a swollen tip at the end of the germinating hyphae (Fig. 3A).

GFP-labeled P. palmivora during colonization at different times in oil palm immature leaflets. A: Appressorium, GC: Germinating cyst, AH: Apoplastic hyphae, HS: Hyphal swelling, H: Haustorium, Sp: Sporangium. The scale bar represents 50 μm.
Subsequently, we observed the first hyphae penetrating and colonizing the apoplast between three and six hours post-inoculation. An increase in apoplastic hyphal growth was also observed, as well as the caducity of the germinating cyst, which was not fluorescent at 6 hpi (Fig. 3B).
Between 12 and 24 h, an abundant presence of mycelium in the apoplast and the first haustorium were visualized (Fig. 3C, D). Finally, at 48 h after inoculation, we detected the necrotrophic phase of the microorganism, represented in new sporangia formation and complete apoplast colonization (Fig. 3E).
Distal parts and rigid immature tissues exhibit cell death responses to counteract P. palmivora colonization
Although there are no completely resistant oil palm materials to bud rot disease, it has been observed that P. palmivora easily infects immature leaf tissues but not mature green tissues (Sarria et al., 2015). Fig. 4A shows that P. palmivora is able to penetrate the tissues of immature oil palm leaflets, which are closer to the rachis from the leaf minus six, and colonize successfully. We also observed that green leaflets from the spear leaf only allowed pathogen colonization in wounded tissues (Fig. 4B). Furthermore, immature leaflets that showed an increased degree of maturity and rigidness also impeded the penetration and colonization of P. palmivora (Fig. 4A). Moreover, Velez et al. (2015) showed that the part of the leaflet proximal to the rachis is more susceptible to P. palmivora than the distal portion.
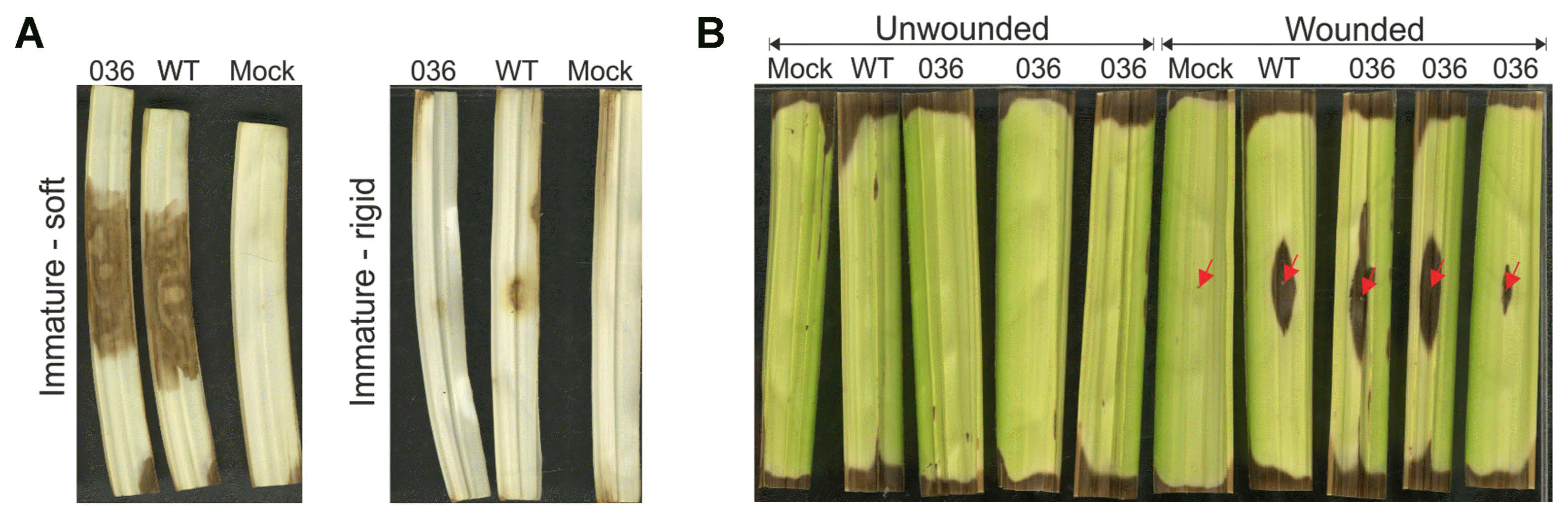
Necrotic lessons observed on immature leaflets inoculated with a WT and GFP-tagged P. palmivora. (A) Soft tissue from minus five leaf and rigid immature tissue from minus three leaf inoculated with P. palmivora, observed at four days post-inoculation. (B) Wounded or unwounded green spear leaflets four days after inoculation with P. palmivora. The red arrow shows the site that were wounded with a sterile syringe needle.
To determine whether P. palmivora is blocked during the penetration or colonization process in less susceptible tissues, we performed an assay to compare the growth pattern of P. palmivora by fluorescence microscopy using eGFP and mRFP1 transformants that were counterstained with PI or FDA, respectively. We obtained neither good quality images with the mRFP1 transformant nor appropriate plant cell staining with FDA (data not shown), and therefore we chose the combination of eGFP - PI to detect P. palmivora structures and to differentiate living from dead plant cells in further experiments.
We inoculated tissue from the proximal part of the youngest soft leaflets, the distal part from a young and soft leaflet and a proximal part of an immature leaflet that showed some rigidness; observations were conducted at 6, 12, 24 and 48 hpi. In the soft young tissue from the proximal part, we observed abundant growth of mycelia but not plant cell nuclei stained with PI, an indicator of cell death (Fig. 5A). In contrast, in the distal part and the rigid tissue, we observed restricted colonization of the apoplastic hyphae and many plant cell nuclei stained with PI (an indicator of cell death), around P. palmivora hyphae, which indicates that cell death responses may be related to the lower susceptibility exhibited in those tissues (Fig. 5C, D). We also observed that some of the cysts were unable to penetrate the distal parts of the immature leaflets (Fig. 5B).

Cell death responses observed in distal and rigid parts of immature leaflets inoculated with a GFP-tagged transformant and counterstained with PI. (A) Proximal soft tissue inoculated at 24 hpi. (B) Germinating cysts of P. palmivora inoculated in a distal part of an immature leaflet at two hpi. (C), (D) Oil palm cell death responses in a rigid immature leaflet at 12 (C) and 48 (D) hpi, respectively. The scale bars represent 100 μm. A: Appressorium, GC: Germinating cyst, PpN: P. palmivora cell nucleus, EgN: Oil palm (E. guineensis) cell nucleus.
Discussion
In this study, we present the successful transformation of three isolates of P. palmivora with different fluorescent proteins, and the respective visualization of the infective process in immature oil palm leaflets. Moreover, we observed that the combination of GFP-transformed isolates with PI staining was useful to elucidate cell death responses in tissues with reduced susceptibility to the infection.
As mentioned previously, P. palmivora has been transformed using three different methodologies, but zoospore electroporation is most commonly used because it does not require enzymes for cell wall degradation or long periods of cocultivation (Huitema et al., 2011). Additionally, the plasmids containing fluorescent protein genes available for oomycete transformation, which were developed by Ah-Fong and Judelson (2011), can only be used for protoplast transformation or electroporation because they do not contain the left and right borders required for ATMT. However, only two isolates of P. palmivora have been transformed using electroporation, a technique that has not been feasible for many Phytophthora strains (Huitema et al., 2011; Rey et al., 2015).
To increase the number of tools available to facilitate experiments of functional genetics of the pathogen, we adapted the methodology developed by Vijn and Govers (2003) by altering the A. tumefaciens strain and increasing the initial amount of zoospores and bacteria during the cocultivation. We also broadened the tools for functional studies of genes in oomycetes by cloning the fluorescent protein genes in the binary vector pNPTII. The resulting binary vectors also have the potential to be used to generate fusion proteins that may be localized to different cellular organelles because they are derived from the plasmids developed by Ah-Fong and Judelson (2011).
Although ATMT has not been widely used in oomycetes (Judelson and Ah-Fong, 2009), it has been shown to be superior to other methodologies in fungi because the production of protoplasts may affect the fitness of the pathogen (Dunn et al., 2013; Keller et al., 1990). Additionally, the probability of single copy transformants is much higher in comparison to other methodologies (Frandsen, 2011). Our results showed that 22 out of 65 transformants were single copy and 33 out of 65 carried two insertions (Table 2); Vijn and Govers (2003) also found that five out of five colonies of P. palmivora and six colonies of P. infestans had a single T-DNA insertion and recently (Wu et al., 2016a) only obtained single copy transformants in their experiments. In plant transformation, it has been widely observed that the use of hypervirulent strains such as AGL1 may have a lower frequency of single copy transformants compared with GV3101 or EHA105 (Chetty et al., 2013). Consequently, we hypothesize that the increase in multiple copy transformants may be affected by the virulence of the A. tumefaciens strain.
Because P. palmivora has a coenocytic mycelium, it is important to check the heritability of DNA fragments inserted into the genome because some hyphae can carry both transformed and untransformed nuclei (Judelson and Ah-Fong, 2009). Zoospores has become the main target structure for oomycete transformation, because are single-nucleated cells that lack a cell wall; however, they may display asynchrony in encystment and elongation of the first hyphae. Thus, it is possible that the cysts may be partially transformed and eventually generate chimeric mycelia. Approximately 40% of the obtained transformants had an abnormal appearance or lost viability during successive passages in selection medium, so we hypothesize that some of them may be a mixture of transformed and untransformed mycelia (Table 2).
Using Southern blotting, we found that the single zoospore cultures had the same number of insertions as the parental cultures, indicating that the T-DNA had been properly integrated into the genome. However, qRT-PCR analysis revealed small variations in the expression levels of fluorescent protein, indicating that colonies derived from different zoospores may have some variations in transgene expression (Supplementary Fig. 1). Vijn and Govers (2003) also, reported that some of the single zoospores derived from transformed cultures lost transgene expression, which may be related to post-transcriptional gene silencing or epigenetic factors.
It is widely known that transformation procedures may affect the growth, virulence or fitness of filamentous plant pathogens (Dunn et al., 2013; Judelson and Ah-Fong, 2009; Wu et al., 2016b). In our research, we tested the in vitro growth and virulence of the brighter transformants for each fluorescent protein. For the in vitro growth assay, we intentionally included the 010 transformant, which has an abnormal aspect and an evident lower growth rate (Table 3). We observed that transformants 021 (CFP-SKL), 032 (eGFP) and 079 (mRFP1) had the same colony diameter and lesion length, indicating that the transformants had the capability to grow and infect oil palm tissues in the same way as the WT isolate. Surprisingly, transformant colony 036 (eGFP) exhibited an increased in vitro growth rate compared with the WT, but the lesion length in the oil palm was not significantly different (Table 3).
The reasons for the differences between the transformants and wild type isolates of filamentous pathogens are not clearly understood, but in vitro colony growth as well as virulence must be evaluated to select transformants that most resemble the WT isolate (Dunn et al., 2013). The introduction of fluorescent proteins has not been clearly correlated to the reduction of growth or virulence of pathogens in oomycetes or fungi because transformant colonies with similar GFP expression may exhibit differences in growth or virulence (Dunn et al., 2013; Si-Ammour et al., 2003; Wu et al., 2016b). Another hypothesis was that random insertion of the T-DNA may alter the ORF of genes related to nutrient uptake or virulence (Frandsen, 2011). In contrast, Wu et al. (2016b) obtained transformants of Fusarium verticillioides with ATMT with a T-DNA insertion in non-coding regions that showed reduced growth compared with the WT at different pH values. Asexual progenies derived from single zoospore cultures of P. infestans exhibited some variations in virulence and sensitivity to Mefenoxam, indicating that even asexual reproduction itself may produce genetic variation between different transformant colonies (Abu-El Samen et al., 2003).
We successfully visualized all P. palmivora structures with conventional fluorescence microscopy, although this was not feasible in previous reports (van West et al., 1999b). However, visualization of the interaction can be greatly improved using modern LSCM. Based on our observations, we visualized the complete infectious cycle of P. palmivora and detected all of the classical structures involved in an infectious process, such as appressoria, germinating cysts, apoplastic hyphae, hyphal swellings, haustoria and sporangia (Fig. 3). We also observed the completion of the cycle in approximately 36 h as previously shown by Sarria et al. (2015) in oil palm, and the transition of the biotrophic phase to the necrotrophic phase, as described in previous reports using LSCM in both monocots and dicots (Le Fevre et al., 2016; Rey et al., 2015).
Mature oil palm tissues, as well as leaflet distal tissues from the rachis, have displayed reduced susceptibility to P. palmivora according to previous works (Sarria et al., 2015; Velez et al., 2015). Using fluorescence microscopy, we confirmed that immature soft tissues that were closer to the rachis were successfully penetrated and colonized by P. palmivora (Fig. 5A). Moreover, we observed that in immature distal tissues from the rachis or rigid immature leaflets, some P. palmivora cysts may form appressoria but are unable to penetrate into the apoplast (Fig. 5B). We also observed that the nuclei of the cysts that did not penetrate were stained with PI, indicating that these structures had lost their viability (Fig. 5B). The hyphae of the cysts that penetrated into the apoplast were associated with plant cell death interactions (Fig. 5C, D). Taken together, these results indicate that oil palm tissues may exhibit age-related penetration resistance. Previous works examining the interaction between grape and Uncinula necator (Ficke et al., 2002), and Nicotiana benthamiana and P. infestans (Shibata et al., 2010) correlated the age of the plant or the maturity of the tissue with an increase in the resistance to an specific disease; however, the molecular basis underlying this characteristic remains poorly understood.
Le Fevre et al. (2016) found that the interaction between barley and P. palmivora displayed a maturity-related susceptibility in terms of both the leaf part and position, and this finding was correlated to Mlo expression. MLO (mildew resistance locus) is a transmembrane protein with unknown biochemical function. However, it is widely known that mlo mutants are resistant to powdery mildew in different plant species, but some may display abnormalities in leaf developmental processes (Reviewed by Acevedo-Garcia et al., 2014). However, more specific studies must be conducted to elucidate the genetic mechanisms involved in plant resistance and the relationship with developmental stages.
The cell death responses observed in our experiments (Fig. 5) resemble the reaction displayed by Arabidopsis thaliana pen2 mutants inoculated with P. infestans. These mutants allowed the penetration of P. infestans, but a secondary mechanism associated with cell death patterns restricted pathogen infection (Lipka et al., 2005). Localized cell death has also been observed in 35-day-old N. benthamiana plants inoculated with P. infestans, but not in 20-day-old plants, supporting the evidence for age-related leaf penetration resistance in the oil palm – P. palmivora pathosystem.
In summary, our study demonstrates the successful transformation of P palmivora isolates with fluorescence proteins using ATMT, and its usefulness to elucidate cellular interactions with oil palm. Moreover, we produce additional evidence to support the presence of age-related resistance in this biological system.
Supplementary Information
Acknowledgments
We thank Dr. Francine Govers at Wageningen University and H. Judelson at UC Riverside, who kindly provided the transformation plasmids, Dr. S. Schornack for his scientific advice during the transformation and real-time PCR experiments, the Pest and diseases group of CENIPALMA for providing the P. palmivora isolates, the Molecular Biology Laboratory technicians D. Guevara, J. Tafur and D. Ramos for their assistance with all experiments, BSc students Gabriel Ortiz and Andrés Parra for performing the single zoospore cultures, Dr. I. Ayala and E. Mesa for their support with the statistical analysis. This work was funded by Fondo de Fomento Palmero administered by Fedepalma, CENIPALMA and COLCIENCIAS.