An in vitro Actinidia Bioassay to Evaluate the Resistance to Pseudomonas syringae pv. actinidiae
Article information
Abstract
Pseudomonas syringae pv. actinidiae (Psa) is by far the most important pathogen of kiwifruit. Sustainable expansion of the kiwifruit industry requires the use of Psa-tolerant or resistant genotypes for the breeding of tolerant cultivars. However, the resistance of most existing kiwifruit cultivars and wild genotypes is poorly understood, and suitable evaluation methods of Psa resistance in Actinidia have not been established. A unique in vitro method to evaluate Psa resistance has been developed with 18 selected Actinidia genotypes. The assay involved debarking and measuring the lesions of cane pieces inoculated with the bacterium in combination with the observation of symptoms such as callus formation, sprouting of buds, and the extent to which Psa invaded xylem. Relative Psa resistance or tolerance was divided into four categories. The division results were consistent with field observations. This is the first report of an in vitro assay capable of large-scale screening of Psa-resistance in Actinidia germplasm with high accuracy and reproducibility. The assay would considerably facilitate the breeding of Psa-resistant cultivars and provide a valuable reference and inspiration for the resistance evaluation of other plants to different pathogens.
Kiwifruit bacterial canker which is caused by Pseudomonas syringae pv. actinidiae (Psa) is so far the most destructive disease of cultivated kiwifruit in the world. The disease can cause leaf spotting, leaf loss, bud browning and drop, fruit desiccation and shriveling, cane dieback and even vine death and lead to great economic losses (Serizawa et al., 1989; Vanneste, 2017). However, there are to date no cures for Psa when once infection has occurred (Donati et al., 2014). All existing treatments are only preventative methods and greatly increase production and management costs. The application of chemical agents may induce resistance in pathogens (Colombi et al., 2017) and cause phytotoxic symptoms to kiwifruit and even damage to the local environment.
Sustainable development of the kiwifruit industry requires Psa-resistant kiwifruit cultivars. However, so far the resistance levels in most existing kiwifruit cultivars or wild genotypes are obscure (Froud et al., 2015). The genus Actinidia has been classified into 55 species and 21 varieties in the latest revision (Li et al., 2007) and most of them originate from China. Nevertheless, nearly all the existing commercially important kiwifruit cultivars are derived from A. chinensis due to its advantages in horticultural and productive traits. Most of these kiwifruit cultivars are susceptible or moderately resistant to Psa (Vanneste et al., 2011).
In 2010, Psa was found in New Zealand and seriously threatened the kiwifruit industry there (Vanneste, 2012). Since then, breeding Psa-tolerant or resistant kiwifruit cultivars has become an important priority for the New Zealand Institute for Plant & Food Research Ltd. Now two new cultivars, ‘Zesy002’ (commonly known as G3) and ‘Zesy003’ (known as G9) that show greater tolerance of Psa than ‘Hort16A’, the highly susceptible cultivar, have been released to growers. The Psa-tolerant cultivar G3 has allowed the New Zealand kiwifruit industry to survive and even expand, and has been proven to be profitable. The identification of genetic resources resistant or tolerant to Psa and their use for breeding Psa-resistant or tolerant cultivars are important for the kiwifruit industry in different countries.
Since 1980, our research group has focused on Actinidia germplasm collection, preservation, and evaluation and established the largest Actinidia germplasm repository in South China. Over the past 30 years, we have identified cultivars or wild genotypes with relatively high resistance to Psa, and we believe that there are more resistant individuals yet to be found.
In the past, the identification of genotypes resistant or susceptible to Psa was mainly under field experiment conditions, but field monitoring can be affected by the pathogen inoculum (Liu et al., 2016), climatic factors (Serizawa and Ichikawa, 1993a; Serizawa et al., 1994), environment (Ferrante and Scortichini, 2014; Vanneste et al., 2015), and vine vigor as influenced by orchard management practices (e.g., water-fertilizer supplies, pruning and the use of chemical agents) (Ferrante et al., 2012). The evaluation of the resistance to Psa through field experiments usually therefore requires large lands, further long-term field trials and even bioassays.
Seedling bioassays are another way to identify genotypes resistant to Psa (Lei et al., 2015; Nardozza et al., 2015) especially by using in vitro seedlings in the laboratory (Wu and Datson, 2015). In comparison to field screening, seedling bioassays can be easily controlled, give consistent results under standardized controlled conditions and better reflect the natural response of the plants to disease. However, in seedling bioassays, it is difficult to maintain individual uniformity between genotypes, usually requires a complicated operational process and occupies large space.
In vitro shoot-bioassays, which have been widely used in the evaluation of the resistance of many fruit trees to fungal pathogens, have been investigated to test for Psa resistance in kiwifruit (Hoyte et al., 2015; Zhang et al., 2014). However, these assays seldom achieved satisfactory results in kiwifruit, and to date there is no general assay procedure involving the type of plant material, incubation time, temperature, humidity, and even the assessment criteria. As a result, conflicting conclusions have been drawn as to the resistance of particular genotypes to Psa (Shi et al., 2014; Yi et al., 2015). A standard methodology for evaluation of Psa resistance in kiwifruit is required.
The New Zealand Institute for Plant & Food Research Ltd. (PFR) developed two bioassays (Hoyte et al., 2015), the “woody stem bioassay” and the “green stem-stab bioassay”, respectively, involving the inoculation of dormant wood stems and soft green stems to characterize and screen resistant genotypes. The aim was the determination of the Psa resistance status of some commercial cultivars and the early identification of Psa-tolerant or resistant genotypes from seedling populations arising from systematic breeding programs. However, the evaluation process of the two bioassays were complex and required the precise measurement of fine differences in lesion size. The two bioassays might potentially give misleading results because the differences in lesion size could be only a few millimeters.
Based on our existing knowledge on the factors of kiwifruit bacterial canker, a unique in vitro bioassay has been developed in our germplasm evaluation program in the study. The resistance levels of different genotypes were classified into four categories according to their response to Psa and the disease severity (mainly represented as lesion length) determined by in vitro assays was highly consistent with field observations. This in vitro bioassay enables the large-scale Psa-resistance evaluation of Actinidia germplasm.
Materials and Methods
Plant materials
All genotypes used in this bioassay (Table 1) were collected from the Actinidia germplasm repository of Guangxi Institute of Botany (GIB), Chinese Academy of Sciences. The highly susceptible red-fleshed cultivar ‘Hongyang’ was used as a control. All the plants were subjected to the same orchard management practices.
Field evaluation
Field data of these genotypes resistant or susceptible to Psa were collected from kiwifruit researchers or planters of different places home and abroad and partial data had been verified in our decades of field observations or laboratory tests. Their resistance or susceptibility to Psa is classified as follows:
Highly Resistant (HR): There is no visible symptom and vines are apparently healthy;
Resistant (R): Disease symptoms occasionally occur on leaves and twigs with minor incidence of disease;
Moderate (M): There are symptoms mainly on canes and shoots, occasionally appearing on trunks and leaders, but the vines generally did not die;
Susceptible (S): Serious symptoms appear on trunks and main branches, usually causing vine wilting and eventual death;
Highly Susceptible (HS): Very sensitive to Psa and the vast majority of vines quickly die.
When various conclusions on the susceptibility or resistance of a certain genotype from different observers or different places were conflicting, we deferred to the “more susceptible” conclusions.
Inoculum preparation and inoculation
All genotypes were inoculated with Psa strain GxL01. GxL01 was isolated from Guangxi Zhuang Autonomous Region of China and is highly infective on Actinidia. It was originally isolated from A. longicarpa leaf spots and further characterized as biovar 3 (Vanneste et al., 2013). The bacteria were cultured for 48 h on LB solid medium at 25°C. Then a single colony was selected and grown in 20 ml LB liquid medium for about 48 h at 25°C under shaking conditions. The final concentration was adjusted to 1 × 109 cfu/ml with sterile water. Before inoculation, cane pieces were surface-sterilized with freshly prepared 70% ethanol, then rinsed twice with sterile distilled water and air-dried before inoculation.
Ten healthy one-year-old canes with a diameter of about 0.8 cm were collected from each genotype during winter pruning and the 30 cm middle part of the canes were used in assays. Each was punched with a hole-punch of 0.5 cm in diameter deeply into the vascular cambium in the middle between two buds. Ten-microlitre bacterial suspension solution was added into the wound. Another ten cane pieces of each genotype punched in the same way but added with sterile distilled water were used as controls for each genotype. The inoculated cane pieces were placed with the wound site uppermost for about 10 min until the bacterial solution was completely absorbed by the wound without any visible solution remaining on the surface. Subsequently, all the inoculated canes were put on a draining board of a seedling tray and the draining board was covered with 2 layers of sterile absorbent paper in advance. The lower tray was filled with sterile water to the bottom of the draining board. Finally, another two layers of sterile absorbent paper were placed over the cane pieces. Control cane pieces were treated in another sterile tray in the same way. The inoculated and control cane pieces were then transferred to a sterile chamber with 90% humidity and at a temperature of 12°C. Incubators were checked every 3 days to make sure that covering paper remained moist. After about 5 weeks, the temperature was increased to 20°C and the incubation was performed for another week. The assay was repeated from 2015 to 2016.
Disease assessment procedure
The cane pieces of the inoculated control ‘Hongyang’ were examined every week to measure the length of lesion browning around the wounds by removing the bark with a paring knife. After 5 weeks of incubation at 12°C and 1 week at 20°C, once the lesion was found to extend to the whole length of cane pieces of ‘Hongyang’, the barks of the cane pieces of all other genotypes were peeled to measure the length of lesion, and the three most seriously affected cane pieces (ALLT, generally with the longest brown lesions) were calculated. And before this, cane pieces had been examined to determine whether any callus was formed at the cut end (usually the basal end, but sometimes both ends) or any bud had sprouted. Then, the three most severe cane pieces were obliquely cut at the wound to determine how deeply the lesion was extended into the wood. The length of the lesion along the cane and other disease symptoms were used to assess the relative Psa resistance.
To test the effects of temperature on disease development, another ten cane pieces of ‘Hongyang’ were inoculated as above but incubated at 4°C in contrast to the treatment at 12°C, and three replicates were performed.
Statistical Analysis
All data was analyzed with SPSS STATISTICS V. 22.0 (IBM Corp.) system. ALLT values were subjected to an analysis of variance (one-way ANOVA), whenever the ANOVA was statistically significant (α < 0.05) for a specific variable. A Tukey-Kramer HSD test was conducted to assess the differences of the means among all genotypes.
Results
Field resistance assessment
The 18 genotypes studied varied in their response to Psa. On the basis of field observations, the genotypes of Eri-1, ‘Huate’, Tet-1 and Mac-1 were classified as HR; ‘Xuxiang’, Hen-3, Let-2 and Gui-1 were classified as R; ‘Zaoxian’, ‘Kuimi’ and ‘Hayward’ were classified as M; ‘Lushanxiang’, ‘Jintao’ and ‘Bruno’ were classified as S; ‘Jinyan’, ‘Jinxia’, ‘Hort16A’ and ‘Hongyang’ were classified as HS (Table 1).
Disease symptoms of in vitro canes
The lesions on ‘Hongyang’ canes extended slowly in the first 2 weeks of incubation at 12°C and then suddenly expanded in 3 weeks after inoculation. After 5 weeks of incubation, the lesions extended along the full length of canes (30 cm) (Fig. 1A). Incubation temperature had a significant impact on lesion extension. When ‘Hongyang’ canes were incubated at 4°C, the average lesion length was no more than 4 cm after 5 weeks of incubation (Fig. 1B).
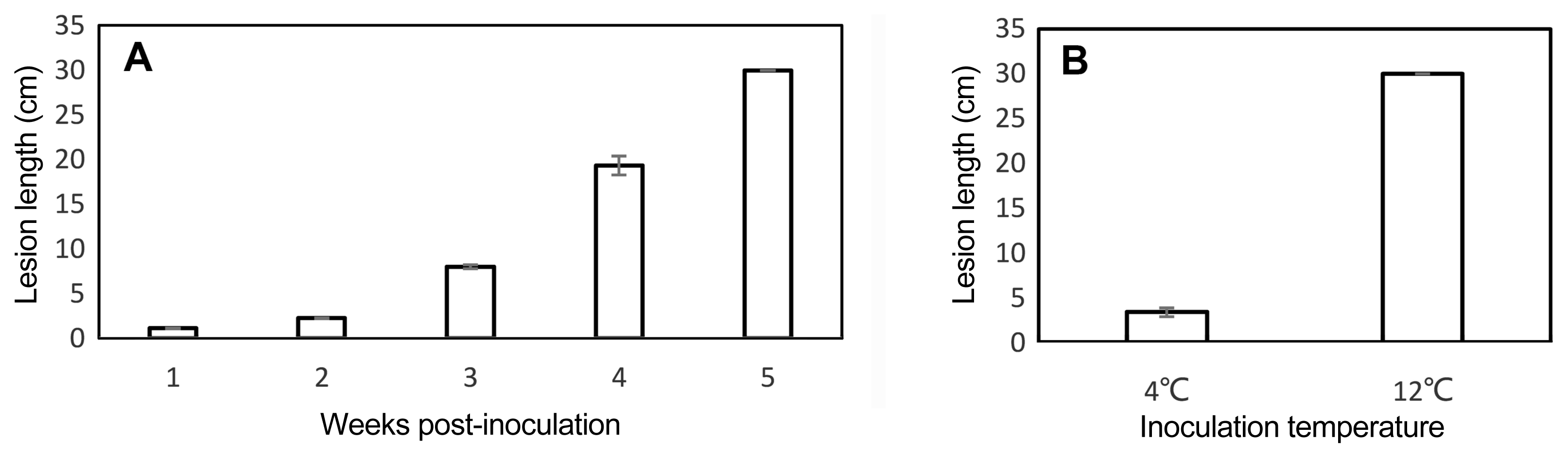
Effects of incubation time and incubation temperature on lesion development of ‘Hongyang’ cane pieces inoculated with a suspension of Pseuodmonas syringae pv. actinidiae. (A) Disease development progress of 1–5 weeks post-inoculation; (B) Effects of incubation temperature on lesion development 5 weeks post-inoculation.
We considered regeneration capacity, characterized as callus formation and bud sprouting, to be a potential indicator of resistance of the plant genotype to disease, because regeneration capacity correlated with resistance in field trials as well as in our experiments. On the canes of all control genotypes that had not been inoculated with Psa, calluses were formed at the cut ends and the buds sprouted after incubation for 6 weeks. Similarly, on the canes of the more resistant genotypes, classified as M to HR, callus formation and buds sprouting were observed (Fig. 2A). As for the genotypes of S, no or few calluses were formed and buds hardly sprouted. As for the genotypes classified as HS on the basis of field observations, callus was not formed and buds did not sprout. Dark-brown Psa ooze came out of the cut ends of the cane pieces, indicating a total collapse of the cane (Fig. 2B).
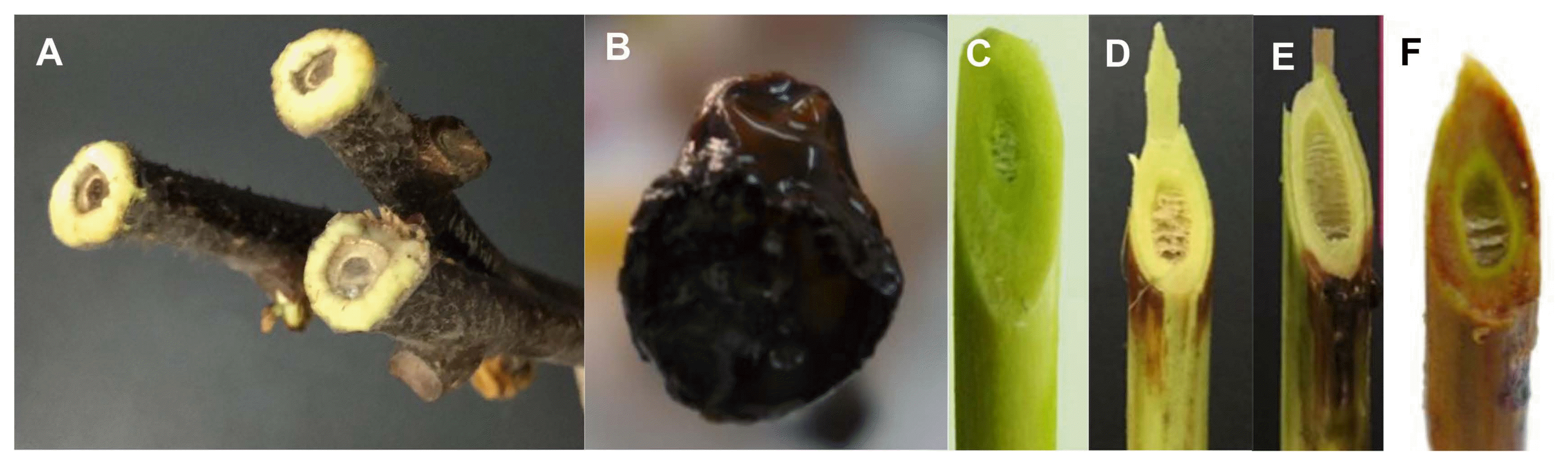
Responses of representative genotypes of different resistant status after inoculation with Pseudomonas syringae pv. actinidiae for 6 weeks. (A) Callus formation and buds sprouting on canes of resistant/tolerant genotype; (B) Dark-brown color on the cane with Psa ooze spilled in highly susceptible genotype; (C) Zero visible lesion symptom on high resistance genotype; (D) Lesion extended only into the outer epidermis and cortex tissues around the wound point for the resistant or moderate genotype; (E) Lesion partly extended into xylem only inside the wound side of susceptible genotype; (F) Lesion completely extended into the entire xylem and eventually the whole cane of highly susceptible genotype turned brown.
After removing the bark, lesions on canes of different genotypes were observed (Fig. 3). There was no visible lesion on the canes of HR genotypes and the canes appeared very healthy. There were only small lesions around the wounds of R genotypes and the lesions were more like dehydrated hypersensitive necrotic tissues that could block the further development of pathogens. Lesions on the canes of M genotypes were also bounded around the wound with minor browning but generally appeared hygrophanous. However, the symptoms of both the formation of dehydrated hypersensitive necrotic tissues in R genotypes and the hygrophanous lesions in M genotypes did not always occur, and sometimes the definite lesion differentiation criteria between R and M genotypes was hard to determine. Compared to the above genotypes, the S and HS genotypes had distinctly extended lesions along the canes and the lesions showed severe browning. Especially, the lesions of HS genotypes covered almost the entire canes, indicating that the entire cane pieces collapsed.

Disease symptoms resulting from the inoculation with a suspension of Pseudomonas syringae pv. actinidiae of cane pieces of 18 genotypes with different levels of resistance. HR, highly resistant; R, resistant; M, moderate; T, tolerant; S, susceptible; HS, highly susceptible; Control is ‘Hongyang’ without the inoculation with Pseudomonas syringae pv. actinidiae.
The length of the lesions formed after the inoculation of cane samples was positively correlated with the genotypic susceptibility, as assessed under field conditions (Table 2). The data showed similarity in repeated tests and ALLT values varied significantly (P ≤ 0.05) between genotypes of different resistance levels through Tukey-Kramer HSD test (Table 2), but no significant difference (P > 0.05) was observed between years (2015 and 2016) using correlation analyses (Table 3).

Disease symptoms of in vitro canes of different Actinida genotypes response to Pseudomonas syringae pv. actinidiae (Psa)
The two years’ mean lesion length of various genotypes showed that: almost the entire length (30 cm) of the canes of the HS genotypes, 8.67–10.42 cm in S genotypes, 3.15–6.87 cm in M genotypes, and 1.93–2.85 cm in R genotypes, and there was no visible lesion, even at the wound, in HR genotypes (Table 2). The extent to which the lesion extended vertically into the wood was also highly correlated with the susceptibility observed in the field. No visible lesion appeared near the inoculation point in HR genotypes (Fig. 2C); lesions extended into only the outer epidermis and cortex tissues around the inoculation point in R and M genotypes (Fig. 2D); lesions partly extended into xylem only on the side of the inoculation point in S genotypes (Fig. 2E); lesions extended throughout the xylem and caused the browning of the whole cane and death in HS genotypes (Fig. 2F).
It should be noted that A. chinensis “Hayward” was classified as “Moderate” according to field observations and other disease symptoms in the assay, but it fell into “Susceptible” classification according to Tukey-Kramer HSD test (Table 2). So it can also be considered to be marginally in the “Susceptible” category to a certain extent.
Classes of resistance to Psa
Various genotypes showed different disease symptoms. HR genotypes had no visible disease damage even at the wound, whereas HS genotypes exhibited the most serious damage with the whole brown cane pieces. Therefore, HR and HS genotypes could be easily distinguished from others. There was no definite lesion difference between R and M genotypes. Although R genotypes seemed more likely to produce dehydrated hypersensitive necrotic tissues at the wound, sometimes the disease severity difference between R and M genotypes was not significant, so the R and M genotypes were collectively referred to as “Tolerant” (T) in this assay (Fig. 3). Compared to R and M genotypes, the S genotypes were generally characterized by more serious lesions along the wound side and into the cane xylem and the limitation of producing callus and breaking buds. Thus the genotypes could be separated into 4 categories according to their disease symptoms (Table 4).

Main disease symptoms used to classify Actinidia susceptibility to Pseudomonas syringae pv. actinidiae (Psa)
As lesion length on canes after incubation was strongly correlated positively with the susceptibility of different genotypes to Psa, it could be used to rank the resistance levels of genotypes within one category. Accordingly, if a higher resolution is required, the symbols of “+” and “−” are used to represent more resistant and more susceptible (+) or less resistant and less susceptible (−) according to the relative lesion length of genotypes in the same category.
Discussion
Psa is so far the most dangerous pathogen of kiwifruit. Infected orchards can be destroyed within two to three years. Previous studies indicated that relatively low temperature (10–18°C) accompanied by high humidity was conducive to Psa propagation and infection (Ferrante and Scortichini, 2014; Serizawa and Ichikawa, 1993a). In order to simulate the conditions favoring Psa, our assays were incubated at a lower temperature of 12°C under the high humidity. Although Gao et al. (2016) reported that Psa migrated faster at 4°C under controlled conditions, the lesion extension on canes of all genotypes was significantly limited at 4°C in this assay (Fig. 1B), which implies that the symptom development on canes may be temperature dependent. It is necessary to increase incubation time: Lesions in ‘Hongyang’ canes initially extended slowly and then lesion length increased rapidly after inoculation for more than 3 weeks (Fig. 1A), implying that the Psa completely prevailed over the host and caused severe disease symptoms. In other reports on in vitro assays (Hoyte et al., 2015; Lei et al., 2015), incubation time of less than 3 weeks were used and lesions had not fully expanded, thus making it difficult to use lesion length to differentiate genotypes of adjacent resistance categories.
Disease symptoms among some kiwifruit cultivars growing in different regions or in different years sometimes vary considerably. A cold and wet climate promoted the outbreak of bacterial canker (Serizawa and Ichikawa, 1993a, 1993b). Such conditions should therefore be used to determine whether a genotype is resistant to Psa. A high humidity, a lower temperature, and a prolonged incubation time were adopted in our assay to simulate such environmental conditions. In this case, pathogens multiplied rapidly, but the physiological activity of the host canes was partially limited with uniformly low vigor. Thus this assay mainly discloses the genetic resistance rather than the physiological sustainability of host plants.
A higher temperature benefits sprouting of the buds and callus formation at the cut ends of the pieces of cane (Serizawa et al., 1994). Tolerant or resistant genotypes were more likely to recover in a favorable environment, and the susceptible ones might perish completely during long-term incubation and lose the capability to produce callus or break buds. Therefore, Psa-tolerant and Psa-susceptible genotypes could be identified. However, it should be noted that certain types of genotypes even being classified as highly resistant levels sometimes failed to sprout due to their long chilling requirements.
Although field monitoring provides direct evidence of the resistance of certain genotypes, the results are not always reliable because Psa causes serious symptoms only under favorable conditions. The responses of a particular genotype in one region does not necessarily indicate the general susceptibility or resistance of that genotype, so the data from field observations should be crosschecked and comprehensively considered. The responses of most of the genotypes to Psa in this assay were the consequence of years of field observations and laboratory tests by orchardists and scientists both from different provinces of China and abroad. However, the conclusions on susceptibility or resistance are sometimes inevitably conflicting. When the conclusions differed, we accepted the “more susceptible” conclusions as probably more realistic. Similarly, we determined the susceptibility based on the three most seriously affected canes in the in vitro assay.
It is also a warning that the experimental approach used can sometimes affect evaluation results and even lead to conflicting conclusions. For example, some researchers generally use the disease index to evaluate the susceptibility of individual genotypes by counting the number of the leaves with disease symptoms, but the approach is not always appropriate for assessing Psa disease severity in kiwifruit. As observed by Nardozza et al. (2015) and Vanneste et al. (2014), the characteristic disease symptoms were found on the leaves of the higher resistant genotypes rather than the susceptible ones in early stages after the inoculation with Psa, suggesting that the highly resistant kiwifruit cultivars might have a complicated and special response to Psa invasion. Thus, the assessment of resistance to Psa simply by observing leaf symptoms might lead to misleading conclusions.
Due to the lack of an accepted in vitro method for the rapid evaluation of Psa resistance in Actinidia, the identification and use of highly resistant cultivars or wild individuals to breed tolerant or resistant scion and rootstock cultivars have been delayed, thus limiting practical solutions to the problem of Psa. By far, only a few cultivars had been proven to be resistant or tolerant to Psa through years of field screening and laboratory tests. By contrast, there seemed to be relatively more number of resistant or tolerant genotypes in wild species. A large-scale monitoring of kiwifruit germplasm collections for Psa resistance or tolerance kiwifruit germplasm was carried out in New Zealand (Nardozza et al., 2015) and revealed promising resistance (or tolerance) in species belonging to the Section Leiocarpae of Actinidia. In our previous study, we also found that some genotypes of other species, such as A. eriantha and A. latifolia, were considerably resistant to Psa. Such species had important breeding characteristics, such as high vitamin C content, easy peeling, high yield, and high resistance to other diseases and insect pests. Many excellent cultivars and wild Actinidia genotypes resistant to Psa may be omitted.
The absence of an effective in vitro method for determining and evaluating resistance of Actinidia genotypes to Psa also restrains the mapping and cloning of Psa-resistance genes by genetic map construction and marker-based association analysis, which are based on the precise identification of phenotypic resistance of recombinant inbred population. The in vitro assay method described here can be readily applied under controlled conditions to evaluate Psa resistance in individual Actinidia genotypes. This is the first report of an in vitro assay capable of large-scale screening of Psa-resistant germplasm. The application of this assay will considerably facilitate the breeding process of Psa-resistant cultivars and promote the isolation and cloning of the Psa-resistance genes. In addition, this assay provides a valuable reference and inspiration for the in vitro resistance evaluation of other plants to diseases.
Acknowledgments
This work was supported by the Fund for Less Developed Regions of the National Natural Science Foundation of China (31560509); Fundamental Research Funds of GXIB (19002); Science and Technology Major Project of Guangxi (AA17204026-2, AA17204097-13, AA17204045-4), the Earmarked Fund for China Agriculture Research System (nycytxgxcxtd-13-1) and the Young Scholar of CAS “Light of West China” Program. We thank Ross Ferguson and Jinhu Wu for critically reading and revising the article.

