Characterization of Antibacterial Strains against Kiwifruit Bacterial Canker Pathogen
Article information
Abstract
Kiwifruit (Actinidia spp.) is an economically important crop and a bacterial canker disease, caused by Pseudomonas syringae pv. actinidiae (Psa), is the most destructive disease in kiwifruit production. Therefore, prevent and control of the disease is a critical issue in kiwifruit industry worldwide. Unfortunately, there is no reliable control methods have been developed. Recently, interest in disease control using microbial agents is growing. However, kiwifruit microbiota and their roles in the disease control is mainly remaining unknown. In this study, we secured bacterial libraries from kiwifruit ecospheres (rhizosphere, endospere, and phyllosphere) and screened reliable biocontrol strains against Psa. As the results, Streptomyces racemochromogenes W1SF4, Streptomyces sp. W3SF9 and S. parvulus KPB2 were selected as anti-Psa agents from the libraries. The strains showed forcible antibacterial activity as well as exceptional colonization ability on rhizosphere or phyllosphere of kiwifruit. Genome analyses of the strains suggested that the strains may produce several anti-Psa secondary metabolites. Our results will contribute to develop biocontrol strains against the kiwifruit canker pathogen and the disease management strategies.
Kiwifruit belongs to the family of Actinidiacea and Southeast Asia is known as the origin of kiwifruit. Since 1970s, kiwifruit has been cultivated and became an economically important crop worldwide. Italy, China, and New Zealand are the major countries of kiwifruit production (Balestra et al., 2009). Kiwifruit contains actinidin hydrolyzing enzymes, high concentration of vitamin C and potassium, which has been known to prevent hypertension in human (Kim et al., 2013). Major cultivation species of kiwifruit are Actinidia deliciosa (green-fleshed) and Actinidia chinensis (yellow-fleshed). Representative cultivars Hayward (A. deliciosa) and Hort16A (A. chinensis) are commonly cultivated worldwide.
Bacterial canker disease caused by Pseudomonas syringae pv. actinidiae (Psa) brings the most severe damage in kiwifruit cultivation. The canker disease presents a few distinctive symptoms such as brown discoloration of buds, brown spots with yellow haloes on leaves, milky or reddish bacterial ooze on infected branches and typical canker on trunk (Renzi et al., 2012). Psa, a member of family Pseudomonadaceae, is a gram-negative bacterium, aerobic, rod-shaped, motile with polar flagella, and has broad temperature adaption from 4 to 20°C (Donati et al., 2014). Psa can be sub-classified as five biovar types (Kisaki et al., 2018). Biovar 1 produces phaseolotoxin and biovar 2 secretes coronatine toxin. Biovar 3, which is known the most powerful virulence biovar, has type III effector system and toxin biosynthesis gene clusters. Biovar 5 shows relatively weak virulence and biovar 6 produces both phaseolotoxin and coronatine toxins (Sawada et al., 2016). However, biovar 5 and 6 were reported only in certain topical area in Japan (Fujikawa and Sawada, 2016). But, Psa may rapidly evolve to a new biovar type (Colombi et al., 2017). For example, Xin et al. (2018) reported that pathogenic Pseudomonas strain evolved from a nonpathogenic P. fluorescens–like ancestor. The nonpathogenic P. fluorescens evolved to a pathogenic Pseudomonas strain owing to habitat environmental circumstance.
Population of plant-associated microbes has been estimated at 106–109 cells/g of rhizosphere soil, 106–107 cells/cm2 of leaves and 105–106 cells/g endosphere tissue (Chebotar et al., 2014; Sébastien et al., 2015). Plant growth–promoting rhizobacteria (PGPR) plays important roles in plant health (Kaur et al., 2016) and PGPR also can enhance the host tolerate against abiotic stresses, nutrient acquisition, and improving root function (Vacheron et al., 2013). Plant secretes rhizodepositions, called root exudates, which are attractant molecules to microbes in bulk soil as chemotaxis and the responsible microbes from bulk soil colonize on the rhizosphere (Hardoim et al., 2008). Certain microbes enter inside root tissue using cell wall degrading enzymes or through cracks and become endophytes. Edaphic factors, rhizodeposits, cell wall features, and host genotype may determine the microbial communities in rhizosphere and endosphere (Bulgarelli et al., 2013). Endophytes migrate through cortex, endodermis, central cylinder and arrive at xylem tissue and then ascending migration using plant transpiration stream (Chi et al., 2005). Phyllosphere is an extreme and irregular habitat to microbes due to fluctuating constraints such as changes of temperature, ultraviolet radiation and unstable relative humidity (Bringel and Couée, 2015). Phyllosphere microbes also have been known as important factors for plant growth (Stone and Jackson, 2016). Thus, more understanding of the microbial communities’ structure and function, which are associated with crop is necessary to improve disease managements in many crops (Bonanomi et al., 2018).
In this study, we collected and constructed kiwifruit microbial libraries from rhizosphere, endosphere, and phyllosphere. With the obtained libraries, we screened and selected core anti-Psa strains. Several key biological properties and genome information were characterized the selected anti-Psa stains.
Materials and Methods
Sample collection
Experimental kiwifruit (cvs. Deliwoong, NHK0013) were designated in the kiwifruit genetic resource center in Namhae, Republic of Korea (34°48′56.8″N 127°55′42.7″E). Samples of rhizosphere soil, sap and pollen were taken from a same tree with three biological replications. Rhizosphere soil samples were collected from surface of rootlet of the tree depths between 25 to 40 cm. Attached rhizosphere soil was brushed lightly into 50-ml tubes. The rhizosphere soil was collected on March 24, 2017, temperature was 9.3 ± 4°C and a relative humidity was 68.8%, respectively. Sap was collected by drilling the second branch and securing the sap into sterilized 5-ml tubes on April 12, temperature was 15.9 ± 6°C and a relative humidity was 28.8%. Pollen was collected on May 15, 2017 (18.5 ± 5°C and a relative humidity of 40.6%). Total 100–150 flower buds of the kiwifruit were picked from each tree and then dried in a dry chamber at 23°C for 4–5 h. Then the pollen was collected in 100 μm of testing sieve (NAVIMRO, Seoul, Korea) by shaking off (Cwynar et al., 1979). All samples were collected with three biological replications.
Isolation and construction of bacteria collection libraries
Pollen samples (0.1 g) were mixed with 9.9 ml of 1× phosphate saline buffered (PBS; 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter final pH 7.4) and sonicated with 35 kHz frequencies for 10 min using an ultrasonic bath (Bandelin electronic GmbH & Co., Berlin, Germany) to detach microbial organisms from pollen. Rhizosphere soil (1 g) or 1 ml of sap was mixed with 9 ml of sterilized water. Three replicates of each rhizosphere soil, sap, and pollen samples were diluted with sterile water from 10−1 to 10−5, spread on 1/5 tryptic soy agar (TSA; 6 g of tryptic soy broth [TSB], 20 g of agar per liter) and then incubated at 27°C for 5 days. Single colony was picked by a sterilized toothpick and transferred into 1/5 TSB (6 g of TSB per liter) in 96-well-plates (SPL Life Science, Seongnam, Korea), incubated at 27°C for 3 days and then stored as glycerol stock (20% final concentration) at −80°C until further use.
Screening anti-Psa strains
Anti-Psa assay was designed using a modified disk diffusion method (Ahn et al., 2009) and the assay conducted three times per a bacterium. As the first screening, Psa was cultured in TSB for 7 days. The bacterial libraries were inoculated on tray plates, which contained 1/5 TSA, using a 96-pin microplate replicator (Boekel Scientific, Feasterville, PA, USA) and incubated at 27°C for 3 days. Psa was mixed with 0.5% sterilized agar solution as final OD600 0.2 and the mixed solution were overlain on the TSA tray plates as the second layer using a 1-ml pipette. The agar was completely hardened at room temperature, incubated at 20°C for 3 days and evaluated inhibition activity to select anti-Psa strains. The second and third screenings were performed with the selected anti-Psa bacteria from 1st screening (cell density was adjusted OD600 0.2) and Psa on 90-mm Petri dishes (SPL Life Science). At the third screening, diameter of Psa inhibition zone was measured and statistical analysis was carried out by Tukey’s HSD test (P < 0.05).
Identification of anti-Psa strains and culture condition
To identify, 16S rRNA and housekeeping genes (atpD and recA) were sequenced with the selected anti-Psa strains. The strains were cultured on MS agar (mannitol soya; 20 g of mannitol, 20 g of soya, 20 g of agar per liter). Genomic DNA was extracted by modified CTAB methods (Wilson, 2001). Polymerase chain reaction (PCR) was performed with amplification primers in Supplementary Table 1. Each 50 μl reaction mixtures contained 20 ng of template DNA, 20 μl of 2× PCR buffer for KOD FX Neo, 0.16 mM dNTPs, 1 μl of forward and reverse primers (10 pmol/μl) and 0.3 μl of KOD FX Neo (1.0 U/μl) with PCR-grade water. PCR reactions were initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at primer-pair-specific temperature for 30 s, extension at 72°C for 1 min and final extension at 72°C for 10 min. The PCR products were checked by electrophoresis and cloned into a T-vector (Promega, Madison, WI, USA) then sequencing was performed by Macrogen Co. (Seoul, Korea).
For spore production, the strains were cultured on ISP4 (10 g of starch, 1 g of K2HPO4, 1 g of MgSO4, 1 g of (NH4)2SO4, 1 g of CaCO3, 1 mg of FeSO4, 1 mg of MgCl2, 1 mg of ZnSO4, 18 g of Bacto agar per liter, final pH 7.2) media at 30°C for 5 days. Mature bacterial colonies on the plates were prepared as glycerol spore stocks. One mL of 20% glycerol was added on the plates and all the spores were rubbed using a sterile cotton stick. The cotton was pushed into a 15-ml syringe and suspension was expelled into the Eppendorf tube by replacing the syringes’ plunger. The glycerol spore stocks were adjusted OD600 0.2 and prepared at −80°C for the further experiments.
Hydrolytic enzymes and plant growth–promoting (PGP) activity assays
Selected anti-Psa strains were assessed three hydrolytic enzyme activities (cellulolytic, chitinolytic, and proteolytic) and six PGP activities (siderophore production, phosphate solubilization, indole-3-acetic acid (IAA) production, nitrogen fixation, attachment, and antibacterial activity) using solid media. Cellulolytic reaction media contained 4 g of KH2PO4, 2 g of CaCl2, 2 g of NH4Cl, 1 g of yeast extract, 1 g of carboxymethyl cellulose, 1 g of MgSO4, 0.05 g of MnSO4, 0.05 g of FeSO4, 18 g of agar per liter (final pH 7–7.4). The chitinolytic reaction media contained 5 g of chitin, 5 g of yeast extract, 0.7 g of KH2PO4, 0.5 g of K2HPO4, 0.3 g of MgSO4, 0.1 g of FeSO4, 0.1 g of NaCl, 18 g of agar per liter and adjusted final pH 6.5–7.0 (Kim et al., 2017). Proteolytic reaction media contained 10 g of skim milk powder, 18 g of agar per liter (Deepthi et al., 2012). Bacterial spore stocks (20 μl) were inoculated with 8-mm diameter paper disks and incubated at 27°C for 5 days. Cellulolytic reaction plates were stained with 0.1% aqueous congo red solution, chitinolytic reaction plates were stained with 1% aqueous congo red solution for 15 min and both were washed completely by 1 M NaCl (Kim et al., 2017). Diameter of clear yellow (cellulolytic, chitinolytic) and transparent zones (proteolytic) were measured with three replicates. Statistical analysis was carried out (Tukey’s HSD test, P < 0.05).
For siderophore producing assay, chrome azurol S (CAS) agar media were prepared with three steps (Louden et al., 2011). First, Blue dye solution was prepared with three solutions. Solution 1 was dissolved with 0.06 g of CAS in 50 ml of ddH2O. Solution 2 was dissolved with 2.7 mg of FeCl3-6H2O in 10 ml of 10 mM HCl. Solution 3 was dissolved with 73 mg of hexadecyltrimethylammonium bromide in 40 ml of ddH2O. Solution 2 (9 ml) was mixed with Solution 1 and then Solution 3. Second, respective mixture solutions were prepared. Minimal media 9 (MM9) salt solution was dissolved with 15 g of KH2PO4, 25 g of NaCl, 50 g of NH4Cl with 500 ml of ddH2O. Glucose stock (20%) was dissolved with 20 g of glucose with 100 ml of ddH2O. NaOH stock was dissolved with 25 g of NaOH in 150 ml of ddH2O as final pH 12. Casamino acid solution was prepared overnight at 4°C with 3 g of casamino acid in 27 ml of ddH2O and 30 ml of 3% 8-hydroxyquinoline in chloroform and filtered sterilize to remove any trace iron. Finally, CAS agar was prepared as follows. One hundred milliliters of MM9 salt solution was added to 750 ml of ddH2O and dissolved with 32.24 g of PIPES (piperazine-N,N′-bis [2-ethanesulfonic acid]). Total 15 g of agar was added, autoclaved and cooled to 50°C. Thirty milliliters of sterilized casamino acid solution and 10 ml of 20% sterilized glucose stock was added. Blue dye solution (100 ml) was slowly mixed along the glass wall and poured in the plates. Bacterial spore stocks (20 μl) were inoculated on the CAS agar media and incubated at 27°C for 5 days.
For phosphate solubilizing assay, Pikovskaya’s medium was prepared (Pikovskaya, 1948). Pikovskaya’s medium contained 10 g of glucose, 0.5 g of (NH4)2SO4, 0.2 g of KCl, 0.1 g of MgSO4, 0.002 g of MnSO4, 0.002 g of FeSO4, 0.5 g of yeast extract, 5 g of Ca3(PO4), 20 g of agar per liter, final pH 6.8–7.0. Inoculated methods of the spore stocks and verified method of clear yellow zones were the same as the enzymes assay. For IAA production assay, Salkowski reagent method was employed (Patel and Parmar, 2013). First, the strains were incubated in tryptophan-containing and free 1/5 PDK broth (2 g of potato dextrose broth, 2 g of Bacto peptone per liter) at 30°C for 14 days as three replicates. Cell was centrifuged at 5,000 rpm for 10 min and the 1 ml of aliquots were transferred into 1.5-ml test tubes. The Salkowski reagent was prepared with 35% of HClO4 and 0.5 M of FeCl3. The aliquots were mixed with 2 ml of Salkowski reagent as 2:1 ratio. The color change to pink was observed for 25 min and measured by spectroscopy at 530 nm. For nitrogen fixation assay, semi solid NFb medium was produced according to Baldani et al. (2014). The NFb medium contained 5 g of malic acid, 0.6 g of K2HPO4, 0.4 g of KH2PO4, 0.01 g of MnSO4, 0.05 g of MgSO4, 0.02 g of NaCl, 0.002 g of Na2MoO4, 2 ml of Bromothymol blue (0.5% in alcohol) and 1.75 g of agar per liter final pH 6.6–7.0. NFb medium (5 ml) in test tubes were autoclaved and cooled. Each of the spore stocks (OD600 0.2) was vertically inoculated in the middle of NFb medium using a loop and incubated at 27°C for 7 days. Attachment abilities were verified using crystal violet dyeing methods with three replicates (De Jong et al., 2009). To evaluate adherent ability, the strains were cultured in 5 ml of minimal NMMP medium (2 g of (NH4)2SO4, 5 g of casamino acids, 0.6 g of MgSO4, 50 g of polyethylene glycol 6000, 1 ml of minor elements solution, 15 mM NaH2PO4, K2HPO4 buffer (pH 6.8), 0.5% glucose per liter) at 27°C for 10 days. Non-adherent cells were removed by washing with 5 ml of 1× PBS. Crystal violet solution (1%, 10 ml) was added to each tube and incubated at room temperature for 3–4 h. All solutions were removed using a pipette and each tube was completely dried at room temperature overnight. Crystal violet was solubilized with 2 ml of 95% high performance liquid chromatography grade ethanol. The turbidity was compared with control sample and OD570 was measured by a spectroscopy. Statistical analyses were evaluated by Tukey’s HSD (P < 0.05) in Statistix (v. 8.0). Sporulation assays were tested on 1/5 PDK agar. The spore stocks (5 μl; OD600 0.2) were inoculated, grown at 30°C and sporulation abilities were daily examined for 7 days.
Intergeneric conjugation to generate antibiotic selection marked stains
For the verification of colonization and Psa inhibition of the selected strains in planta, intergeneric conjugation between Escherichia coli and the selected Streptomyces sp. strains was performed as Paranthaman and Dharmalingam (2003). E. coli ET12567 (pUZ8002/pIJ10257) and E. coli ET12567 (pUZ8002/pSET152) were grown to OD600 0.4 in 10-ml LB broth (25 g of Luria-Bertani per 1 liter). E. coli was obtained by centrifugation, washed twice with fresh LB broth (10 ml) and resuspended in 500 μl of LB broth. Glycerol spore stocks of Streptomyces sp. (10 μl, OD600 1.0) were resuspended in 500 μl of 2× YET medium (yeast extract tryptone; 10 g of yeast extract, 16 g of tryptone and 5 g of NaCl per liter, final pH 6.8) and incubated at 50°C for 10 min to derive germination. Equal volume (500 μl) of the donor cells and the treated spore stocks were mixed, spread on an MS agar plate and incubated at 30°C for 16 h. The plates were overlaid with 1 ml of sterilized water containing 30 μg/ml of nalidixic acid and 80 μg/ml of hygromycin or 50 μg/ml of apramycin. The plates were incubated at 30°C for 3 days. Several colonies were picked and cultured on the MS agar containing 30 μg/ml of nalidixic acid and 80 μg/ml of hygromycin or 50 μg/ml of apramycin. Respective antibiotic resistance colonies were confirmed, and the colony was prepared as spore stocks for further studies.
Colonization and disease suppressive activity of the anti-Psa strains
Rhizosphere colonization assays were performed using apramycin resistant W1SF4 and hygromycin resistant W3SF9 strains. Bacterial cell densities were adjusted as 106 colony-forming unit (cfu)/ml in 0.1% Tween 20 solution. Roots of kiwifruit seedlings (cv. Deliwoong, 12-week-old) were washed with tap water and the dipped in the bacterial suspension for 15 min. Untreated controls were used as the roots were dipped in sterilized 0.1% Tween 20 solution. Experimental plants were planted in a deepots (Stuewe & Sons, Inc., Tangent, OR, USA) containing garden soil. The plants were maintained in growth chamber at 25°C, 80% humidity, 12 h light photoperiod. Root colonization was measured by diluting the rhizosphere soils from 10−1 to 10−5, spreading on 1/5 PDK agar with selective antibiotics and verifying the cfu/g at weekly intervals up to 6 weeks.
Leaf colonization assays were performed using apramycin resistant W1SF4, hygromycin resistant W3SF9 and KPB2. Each bacterial cell density was adjusted at 107 cfu/ml with 10 mM MgCl2 and 0.02% silwet L-77. Kiwifruits were planted with garden soil in the deepots and the bacterial suspensions were sprayed evenly three times with 2-day intervals onto both adaxial and abaxial sides of the leaves. Leaf colonization was measured by diluting leaves from 10−1 to 10−7, spreading on 1/5 PDK agar with proper antibiotics and verifying the cfu/g. The deepots were maintained for 21 days and evaluated the bacterial strains colonization on leave.
For canker disease suppression assay, Psa (107 cfu/ml) was sprayed on the leaves. The treated plants were maintained for 21 days. The disease index (DI; 0–5) was determined by observing the percentage of diseased leaves; 0 = no symptoms; 1 = trace to 5%; 2 = 5–15%; 3 = 15–30%; 4 = 35–70%; 5 = more than 70%. Statistical analysis was carried out in Statistix (v. 8.0) by Tukey’s HSD test (P < 0.05).
Whole-genome sequencing of W1SF4 and KPB2 strains
Genome sequencings of Streptomyces racemochromogenes W1SF4 (rhizosphere strain) and Streptomyces parvulus KPB2 (pollen strain) were performed using the PacBio (Pacific Biosciences) RSII platform (English et al., 2012). Single colony of the strains was cultured in PDK broth at 30°C for 5 days. Cells were harvested by centrifugation at 8,000 rpm for 10 min. Genomic DNA was extracted using the MG genomic DNA purification kit (Doctor Protein, Seoul, Korea). Concentration of genomic DNA was measured by NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and PicoGreen for screening quality control (QC) criteria. Genomic DNA (8 μg) was used for library preparation of 20-kb SMRTbell templates. The genomic DNA less than 17 kb was determined the actual size distribution by Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The proper genomic DNA greater than 40 kb was purified using AMPurePB magnetic beads (Beckman Coulter Inc., Brea, CA, USA). Total 10 μl library was prepared using PacBioDNA template prep kit 1.0 (Thermo Fisher Scientific). SMRTbell templates were annealed using the PacBio DNA/Polymerase binding kit P6 and the 8 SMRT cells were sequenced using the PacBio DNA sequencing kit 4.0. SMRT cells using C4 chemistry and 240-min movies were captured for each SMRT cell using the PacBio RS II platform by Macrogen Co. Gene functions were assigned by Prokka (v 1.12b) after whole genome was assembled. Genome annotation was conducted by RAST version 2.0 (http://rast.nmpdr.org), antiSMASH bacterial version (https://antismash.secondarymetabolites.org/#!/start) for secondary metabolite analysis and NCBI (http://www.ncbi.nlm.nih.gov) for the accurate gene identification (Aziz et al., 2008; Blin et al., 2018).
Results
Library construction and screen, characterization of anti-Psa strains
Culturable bacteria were isolated from kiwifruit ecospheres (rhizosphere, sap, and pollen), total bacteria densities were higher rhizosphere, pollen and sap in order, 109 cfu/g in rhizosphere soil, 107 cfu/g in pollen and 106 cfu/ml in sap, respectively. Of 1,175 bacterial isolates from rhizosphere, 768 isolates from sap and 1,080 isolates from pollen were collected as kiwifruit-associate bacterial library and tested anti-Psa activity (Supplementary Table 2). After the first anti-Psa screening, 56 isolates from rhizosphere, 156 isolates from sap and 106 isolates from pollen were selected. At second screening, 15 isolates from rhizosphere, 35 isolates from sap and two isolates from pollen showed anti-Psa activity. At the final third screening total 11 isolates (9 from rhizosphere, 2 from pollen) were selected as potential anti-Psa microbial agents. The 11 anti-Psa isolates were named accordingly to kiwifruit cultivar, replication number, isolated tissue origin, and location on library; W1SF3, W1SF4, W1SG3, W2SB7, W3SF9, K1SF1, K2SC9, K2SG8, K3SG10, KPB2, and KPP2. First letter W or K indicated kiwifruit cultivar Deliwoong or NHK0013. Number in second place indicated number of replicates. Third letter S or P indicated rhizosphere soil or pollen. Final alphabet and number indicated the storage location in 96-well plates. W1SF4 and W3SF9, both strains were isolated from rhizosphere, showed the largest Psa inhibition zones as 3.53 cm and 3.92 cm. Other strains ranged inhibition zones from 1.34 to 1.96 cm (Fig. 1).
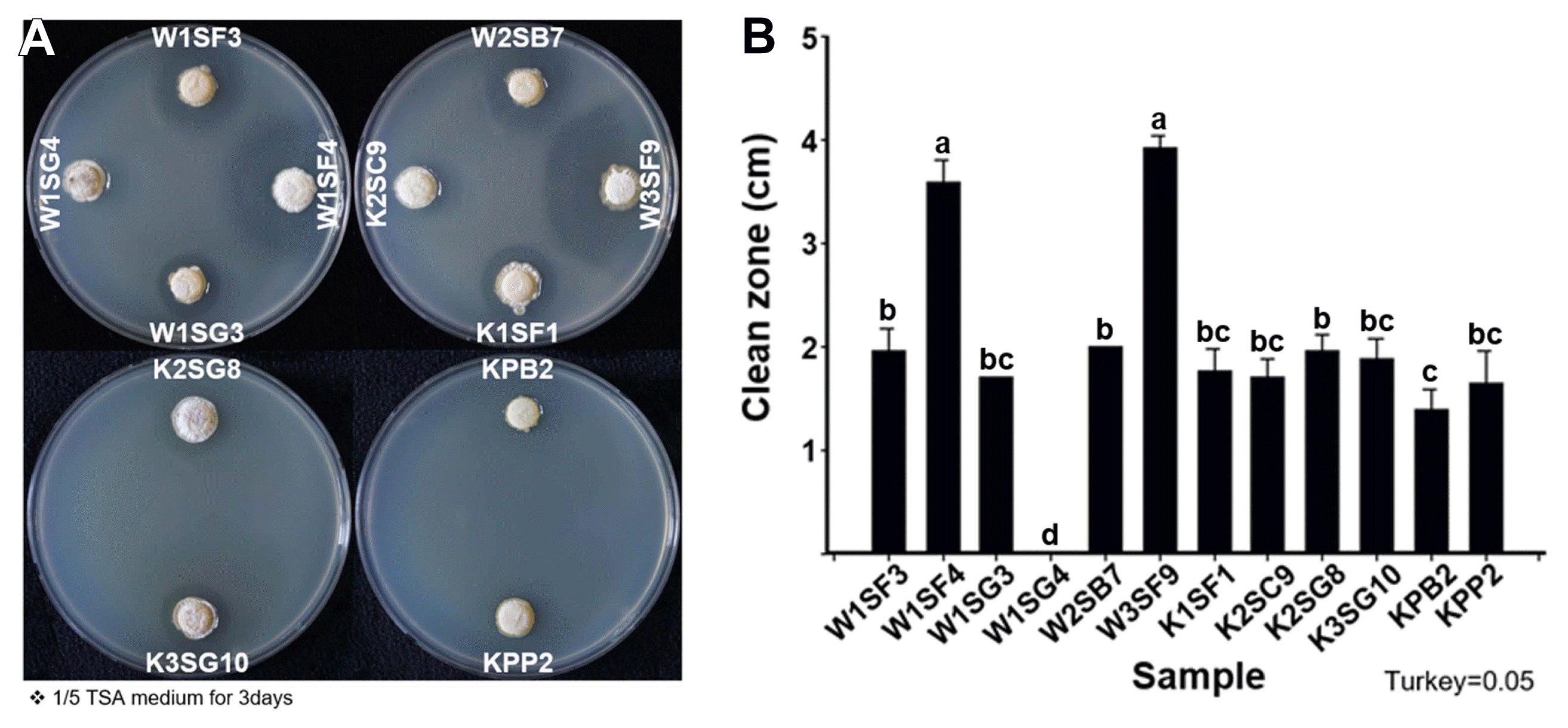
Antibacterial activity of strains against Pseudomonas syringae pv. actinidiae (Psa) by disk diffusion method. (A) 3rd screening plates. (B) Clean zone diameter of selected anti-Psa strains. Co-culture was performed on 1/5 tryptic soy agar (TSA) as OD600 0.2 and incubated at 20°C for 3–5 days. Statistical analysis was carried out in Statistix (v. 8.0) by Tukey’s honestly significant difference test with significance for P < 0.05 (mean ± SD).
All 11 anti-Psa strains were grown and sporulated on MS media. Phenotypic characteristic of the strains was distinguished as six types according to colony color, shape and color change around the colony (Supplementary Fig. 1). Type A produced powdery and less sporulation on white colony with ocher halo, type B presented powdery and abundant light pink colony, type C showed powdery and rough-surfaced white colony with brown halo and type D produced powdery and flat light pink colony. Type E showed powdery and smooth pink colony and type F produced powdery and smooth dusky pink colony. W1SF4, W3SF9, K2SC9, and K2SG8 were grouped as type A. KPB2 and KPP2 were belonged to type F. For type B (W1SF4), C (W3SF9), D (K2SC9), and E (K2SG8), a single strain for each type was assigned (Supplementary Fig. 1).
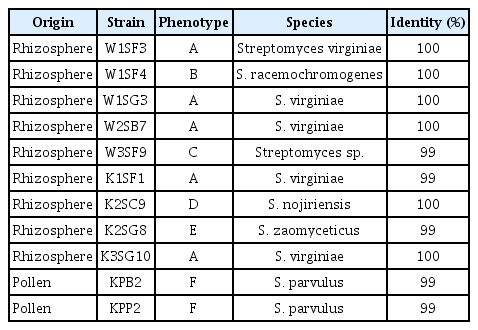
Sequence of 16S rRNA of the strains was compared with references strains in GenBank and all the strains showed 100% similarity with various Streptomyces species but fall to identified in species level. However, the strains can be identified in species level with atpD and recA sequences except W3SF9 (Table 1). The strains showed 98% to 100% similarity with reference strains and identified into species level according to their colony phenotypic types except the type C stain. The type A (W1SF4, W3SF9, K2SC9, and K2SG8) strains were identified as Streptomyces virginiae, B type was S. racemochromogenes, C type was Streptomyces sp., D type was Streptomyces nojiriensis, E type was Streptomyces zaomyceticus and F type was S. parvulus (Fig. 2).
Comparison of anti-Psa strains obtained with housekeeping genes (atpD/recA) sequencing above 98% identity

Phylogenetic tree of the anti–Pseudomonas syringae pv. actinidiae strains based on housekeeping genes atpD (A) and recA (B) by the neighbor-joining method. The sequences of reference species were obtained from NCBI GenBank. The distances were computed using the Maximum Composite Likelihood method and analyses were conducted in MEGA 7.0.26.
In vitro PGP activities of the anti-Psa stain
The selected anti-Psa strains’ cellulolytic, chitinolytic, proteolytic, siderophore producing and antibiotic activities were evaluated (Table 2). Most of the strains showed cellulolytic, chitinolytic, proteolytic and siderophore production activities. However, S. virginiae (W1SF3, W1SG3, W2SB7, K1SF1, and K3SG10) and S. nojiriensis K2SC9 were not shown the proteolytic activity. S. zaomyceticus K2SG8 showed the accomplished proteolytic activity but not presented the chitinolytic and siderophore producing activities. S. racemochromogenes W1SF4, Streptomyces sp. W3SF9 and S. parvulus KPB2, KPP2 had outstanding effects for the cellulolytic, chitinolytic, proteolytic and siderophore producing activities. Prominently, Streptomyces sp. W3SF9 had accomplished antibacterial activity against Psa. Sporulation of S. racemochromogenes W1SF4, Streptomyces sp. W3SF9, S. nojiriensis K2SC9, and S. zaomyceticus K2SG8 were 30% in 3 days and 100% in 7 days, respectively. The sporulation of S. parvulus strains KPB2, KPP2 had shown only about 20% and S. virginiae strains W1SF3, W1SG3, W2SB7, K1SF1, K3SG10 were not sporulated at all for 7 days (Table 2). Through enzymatic and PGP abilities, S. racemochromogenes W1SF4, S. virginiae W2SB7, Streptomyces sp. W3SF9 from rhizosphere and S. parvulus KPB2 from pollen were condensed as anti-Psa strains. S. racemochromogenes W1SF4 had not only the anti-Psa activities but also had PGP and sporulation activities. S. virginiae W2SB7 had antibiotic and PGP activities but no sporulation ability. Streptomyces sp. W3SF9 had anti-Psa and PGP activities. S. parvulus KPB2 was the only species selected from pollen and had effects in all the antibiotics, PGP and sporulation (Table 2).
PGP activities of the strains
These 4 (W1SF4, W2SB7, W3SF9, and KPB2) strains were evaluated additional PGP assays. The strains showed phosphate solubilization, IAA production and nitrogen-fixing activities (Table 2). The four strains also demonstrated attachment abilities, but W2SB7 produced less spore. S. virginiae W2SB7 had several activities but was excluded due to less proteolytic activity and sporulation activity. Among the four strains, S. parvulus KPB2 was only one species from pollen. The three strains (S. racemochromogenes W1SF4, Streptomyces sp. W3SF9, and S. parvulus KPB2) were selected as probiotics for kiwifruit after evaluated various criteria, which were presented above.
Colonization and canker disease suppressiveness of the strains in planta
Density of the strains on root and leaf was measured by plate counts with a proper selection antibiotic maker. In kiwifruit rhizosphere, strains W1SF4 and W3SF9 (rhizosphere strains) colonized at a density of 107 cfu/g of soil up to 6 weeks (Fig. 3A). In phyllosphere, S. racemochromogenes W1SF4 colonized at a density of 106 cfu/g, Streptomyces sp. W3SF9 were detected at a density of 105 cfu/g and S. parvulus KPB2 (pollen strain) presented at a density of 108 cfu/g on the kiwifruit leaves. The canker disease suppression abilities were also verified by disease symptoms and DI. Psa infected leaves were formed yellow or brown hallow as the disease symptoms. The symptoms were shown all areas of leaves on Psa treatments but minor or no symptom was observed on the anti-Psa strain treatments leaves (Fig. 4A). The Psa pathogen only treatment showed near DI 5 at 3-weeks after treatment, but all kiwifruit probiotics strains (W1SF4, W3SF9, and KPB2) significantly reduced the disease severity as under DI 1 (Fig. 4B).
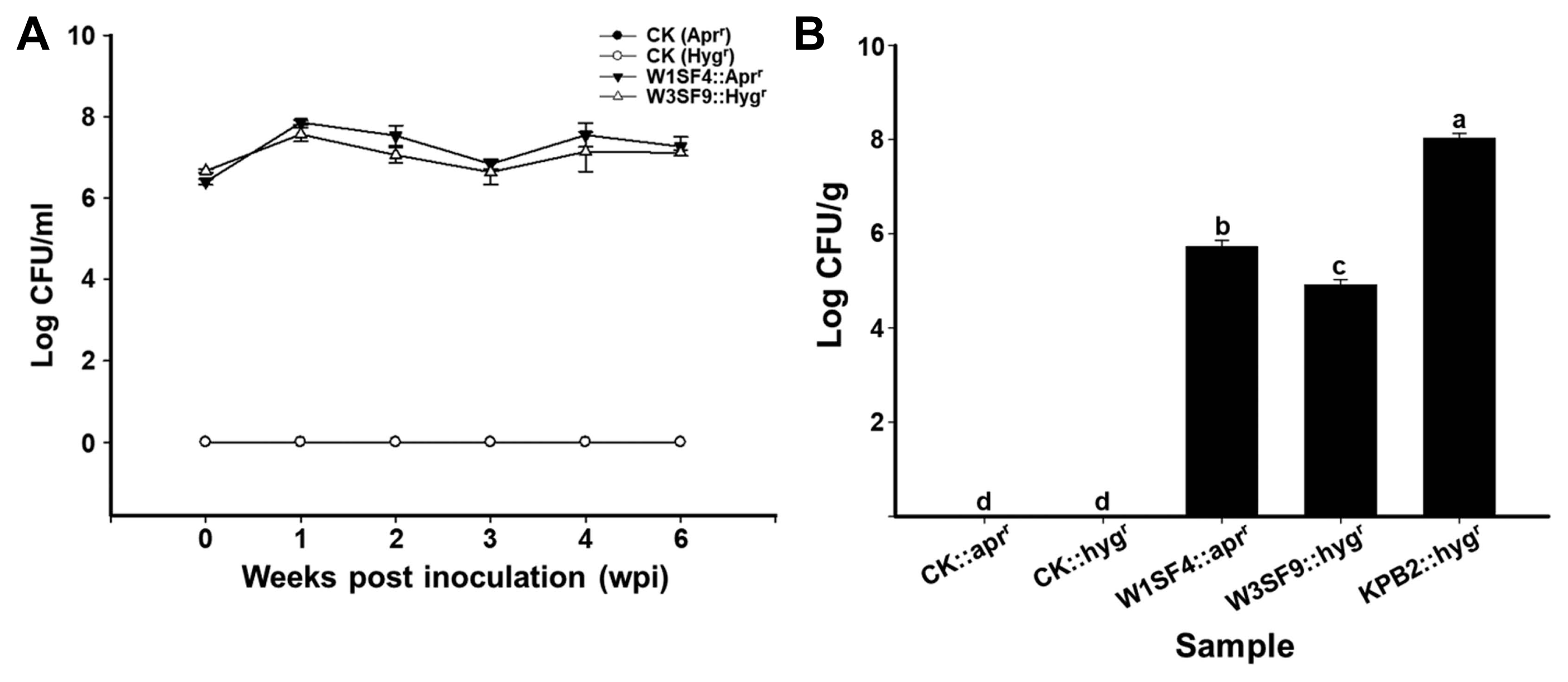
Root (A) and leaf (B) colonization assay of the anti–Pseudomonas syringae pv. actinidiae (Psa) strains. Colonization assays were performed as non-treated control (CK), W1SF4, W3SF9, and KPB2 treatments with five replicates. The anti-Psa treatments were performed with apramycin resistant W1SF4 and hygromycin resistant W3SF9, KPB2. Root colonization assay was performed with bacterial dipping method. Anti-Psa strains were prepared at 106 cfu/ml with 0.1% Tween 20, washed kiwifruit seedlings roots were dipped in bacterial cells for 15 min and planted with sterile bed soil in deepots. The root treatments were maintained in the growth chamber at 25°C, 80% humidity, 12-h light photoperiod. Leaf colonization assay was performed with bacterial spraying method. Each bacterial culture was adjusted at 107 cfu/ml with 10 mM MgCl2 and 0.02% silwet L-77. The kiwifruit seedlings were planted with sterile bed soil in deepots and bacterial suspensions were sprayed evenly three times at 2 days intervals onto both sides of the leaves. The leaf treatments were covered with the lid and maintained on the lab table at from 20 to 25°C for 21 days. The root colonization ability was measured by diluting rhizosphere soils from 10−1 to 10−5, spreading on 1/5 PDK agar and verifying the cfu/g at weekly intervals for 6 weeks. The leaf colonization ability was measured by diluting leaves from 10−1 to 10−7, spreading on 1/5 PDK agar and verifying the cfu/g after a week. Statistical analysis was carried out in Statistix (v. 8.0) by Tukey’s honestly significant difference test (P < 0.05).

The effect of anti–Pseudomonas syringae pv. actinidiae (Psa) strains on the disease severity of kiwifruit bacterial canker. (A) Symptoms on upper surfaces 3 weeks post-inoculation with Psa. Scale bar = 1 cm. (B) Disease index based on the percentage of leaf area covered by symptom. The culture of Psa (107 cfu/ml) was sprayed on the bacterial treatments with the same bacterial spraying methods. They were maintained on the lab table at from 20 to 25°C for 21 days. Statistical analysis was carried out in Statistix (v. 8.0) by Tukey’s honestly significant difference test (P < 0.05).
Complete genome information of W1SF4 and KPB2
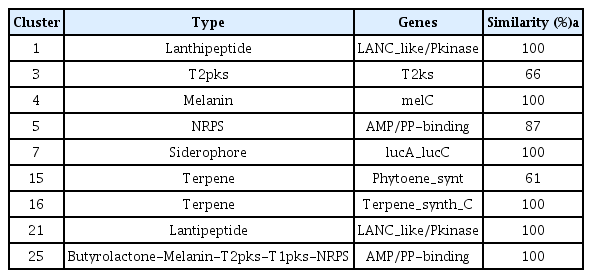
Accession numbers of genome analyses for W1SF4 and KPB2 were presented in Supplementary Table 3. The genome of Streptomyces sp. W1SF4 comprised three contigs of 7,272,878 bp, 481,785 bp and 313,480 bp (Table 3, Fig. 5A-C). Contig 1 predicted 6,899 coding sequences (CDS), contig 2 for 505 CDS and contig 3 for 189 CDS. The GC contents of these components were 73.1%, 72.9%, and 74.5% respectively. The genome carried 216 tRNA and 63 rRNA. Total 25 putative antibiotic secondary metabolite gene clusters were predicted by antiSMASH. Similarity was above 60%, two lanthipeptides, one polyketide synthase (PKS), one melanin, two nonribosomal peptide synthetase (NRPS) and one siderophore, two terpenes biosynthesis gene clusters were presented in Table 4. Genome of Streptomyces sp. KPB2 comprised single linear chromosome of 8,082,236 bp and 7,362 predicted CDS (Table 3, Fig. 5D). This strain carried 54 tRNA, 196 rRNA and GC contents was 72.2%. Total 22 antibiotic gene clusters were predicted and major gene clusters were three NRPSs, three terpenes, two PKSs, one siderophore, one melanin and one ectoine (Table 5). Gene ontology information of the two strains was also presented in Supplementary Table 4 and 5.

Genome map of Streptomyces racemochromogenes W1SF4 and S. parvulus KPB2. (A) W1SF4 chromosome. (B) Plasmid 1 of W1SF4. (C) Plasmid 2 of W1SF4. (D) KPB2 chromosome. The genome map was drawn by annotation result in RAST v 2.0. Marked characteristics are shown from outside to the center; coding sequence (CDS) on forward strand, CDS on reverse strand, GC content and GC skew. The genome map was visualized in R program (v 3.5.1).
Discussion
Bacterial canker disease in kiwifruit, caused by Psa, has tremendous damage on kiwifruit production worldwide. To overcome the disease issue, various methods are being studied such as breeding of resistant cultivar, developing new bactericides, and improving cultivation methods. Unfortunately, there is no promising solution available yet. Problem of the bacterial canker in kiwifruit may not be solved by a single method, it should be applied integrated disease control methods. Therefore, we screened potential anti-Psa biological control agents from kiwifruit bacterial libraries, which was collected from root (rhizosphere), sap (endosphere), and pollen (phyllosphere). Among the libraries, six strains of Streptomyces virginiae, S. racemochromogenes, Streptomyces sp., S. nojiriensis, S. zaomyceticus, and S. parvulus were identified as reliable anti-Psa strains. These strains are mostly reported as having the antibacterial activities. Virginiamycin M/S (VM and VS) of S. virginiae, streptothricin D/E of S. nojiriensis, zaomycin of S. zaomyceticus and Borrelidin (bor) of S. parvulus were identified as antibacterial secondary metabolite agents (Miyashiro et al., 1983; Olano et al., 2004; Pulsawat et al., 2007; Wang et al., 2013). S. racemochromogenes has not been reported antibacterial activity yet, but well understood on transphosphatidylation activity (Nakazawa et al., 2011). After considering element of anti-Psa and PGP activities, S. racemochromogenes W1SF4, Streptomyces sp. W3SF9 and S. parvulus KPB2 were finally selected for further experiments. To be a successful biological control agent, colonization up to 105 cfu/g and maintain population dentistry for certain period on plant tissue is the most critical and essential factor (Cha et al., 2016). These strains (W1SF4, W3SF9, and KPB2) showed superior colonization ability on both the root and the leaf of kiwifruit seedlings. Interestingly, KPB2 (isolated from pollen) colonized at 108 cfu/g of leaf tissue, whereas W1SF4 and W3SF9 (isolated from rhizosphere) were at 105 cfu/g. The two rhizosphere strains presented outstanding colonization ability in root up to 6 weeks. These findings suggested that KPB2 may be more suitable to prevent leaf or above-ground infection of Psa and W1SF4 and W3SF9 can be developed as biopesticide to suppress soil-borne Psa.
To obtain secondary metabolites information in the anti-Psa strains, S. racemochromogenes W1SF4 and S. parvulus KPB2, whole-genome sequencing was analyzed. S. parvulus KPB2 was correctly identified as one linear chromosome DNA, while S. racemochromogenes W1SF4 has three contigs, as a chromosome with two plasmids. S. racemochromogenes W1SF4 was presumed to be a new species because the genome coverage showed less than 70% similarity with a proximate strain in NCBI GenBank. The finding suggested that kiwifruit-associate microbiota has many potentials to discover novel microbes, which may have significant effect on plant health and crop management. Both S. racemochromogenes W1SF4 and S. parvulus KPB2 had typical antibiotic gene clusters such as lantipeptide, PKSs, and NRPS. Currently, streptomycin is the major bactericide to control of kiwifruit canker disease. Yang et al. (2015) report that total 102 Psa isolates were evaluated sensitivity against streptomycin, which inhibited Psa growth at concentration 0.03–0.42 μg/ml. Streptomycin is one of the well-characterized antibacterial antibiotics and its biosynthesis process has been characterized by genome mining (Ohnishi et al., 2008). Interestingly, W1SF4 and KPB2 anti-Psa strains in this study did not have streptomycin biosynthesis gene cluster in their genome. Result of genome mining suggested that the two strains may have different antibiotic mechanisms, for example lantipeptide and its related molecules probably are core metabolites in W1SF4 (Table 4). However, KPB2 does not produce lantipeptie, but the strain may produce variety of NRPS and PKS type antibiotic compounds (Table 5). Further studies, such as knock out mutant or overexpression of the gene cluster, are needed to determine the gene functions, which relates to the antibiotic and PGP abilities for biocontrol products in the field. The anti-Psa strains can be utilized as biocontrol agents in artificial pollination. In addition, antibiotic or PGP compounds can be developed through further individual gene function and metabolite studies.
Supplemental Materials
Acknowledgments
This research was supported by the Next-Generation Bio-Green 21 Program (PJ013250) and Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea (Project No. 315004-5).
Notes
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).