 |
 |
| Plant Pathol J > Volume 40(1); 2024 > Article |
|
Abstract
Garlic can be infected by a variety of viruses, but mixed infections with leek yellow stripe virus, onion yellow dwarf virus, and allexiviruses are the most damaging, so an easy, inexpensive on-site method to simultaneously detect at least these three viruses with a certain degree of accuracy is needed to produce virus-free plants. The most common laboratory method for diagnosis is multiplex reverse transcription polymerase chain reaction (RT-PCR). However, allexiviruses are highly diverse even within the same species, making it difficult to design universal PCR primers for all garlic-growing regions in the world. To solve this problem, we developed an inexpensive on-site detection system for the three garlic viruses that uses a commercial mobile PCR device and a compact electrophoresis system with a blue light. In this system, virus-specific bands generated by electrophoresis can be identified by eye in real time because the PCR products are labeled with a fluorescent dye, FITC. Because the electrophoresis step might eventually be replaced with a lateral flow assay (LFA), we also demonstrated that a uniplex LFA can be used for virus detection; however, multiplexing and a significant cost reduction are needed before it can be used for on-site detection.
Garlic is often infected by many viruses, but garlic yield and quality are mainly affected by mixed infections with leek yellow stripe virus (LYSV), onion yellow dwarf virus (OYDV), and allexiviruses (Jayasinghe et al., 2021; Sasaki et al., 2022; Yoshida et al., 2011, 2018). Thus, periodic testing for viruses is necessary because, although virus-free garlic bulbs can be introduced, garlic plants quickly become infected with viruses in the field. In particular, monitoring for allexiviruses that infect garlic plants in the field is difficult. Seven species of allexiviruses have been currently reported and are characterized by relatively low homology even among isolates of the same species (Kim et al, 2023). In the laboratory, these garlic viruses are mainly diagnosed using an enzyme-linked immunosorbent assay or reverse transcription polymerase chain reaction (RT-PCR) for each virus; for simultaneous detection of the three viruses (LYSV, OYDV, and allexiviruses), multiplex RT-PCR has been reported (Hu et al., 2015; Majumder and Baranwal, 2014; Nam et al., 2015; Park et al., 2005). However, the nucleotide sequence diversity of the viruses infecting garlic in different regions is considerable (Kawakubo et al., 2023; Kim et al., 2023). Although allexiviruses have been detected by RT-PCR in some reports (Chen et al., 2004), often they cannot be similarly detected in garlic plants in other countries using the conditions described (Kim et al., 2023). Consequently, the nucleotide sequences of isolates in the region need to be checked, and the conditions for the multiplex RT-PCR designed for each isolate should be determined. Thus, a portable method that is highly flexible to detect and diagnose the various isolates is needed for laboratory and field use.
We here report the development of a very practical, inexpensive diagnostic method for garlic viruses that is easy to use without specialized knowledge. The method consists of two steps: the first step, an on-site FITC-detection assay (FDA), is used to determine the infection status of garlic plants by detecting the presence or absence of the viruses at the earliest possible time; in the case of a positive finding in the FDA, the second step, an array assay using nucleic acid hybridization is implemented to accurately diagnose the viruses.
Our laboratory group has been developing using array assays to detect plant pathogens such as viruses and fungi for a long time (Furuta et al., 2017; Shimura et al., 2015; Sugiyama et al., 2008). Here, we developed the FDA-array method specifically to detect garlic viruses. However, the method is also broadly applicable for diagnosing other viruses. The usefulness of this method is explained in this study, in comparison to the multiplex RT-PCR method widely used in the laboratory, the rapid immunofilter paper assay (RIPA) method traditionally used for on-site diagnosis of plant viruses, the recombinase polymerase amplification (RPA)-lateral flow assay (LFA) method that has recently begun to be used to detect plant viruses (Kim et al., 2022a, 2022b), and the loop-mediated isothermal amplification (LAMP) method, which is superior due to its high sensitivity (Zhang et al., 2018 for multiplex LAMP).
Leaves were randomly collected from 20 garlic plants in the fields of Tokachi area, Hokkaido. For positive controls, Chinese and Spanish garlic bulbs were purchased at the market.
Total RNA was extracted from garlic leaf tissues or bulbs essentially as described by Yaffe et al. (2012). Garlic tissues (~100 mg) were placed in 1.5 ml microtubes containing 500 ╬╝l RNA extraction buffer (8 M guanidine hydrochloride, 20 mM MES hydrate, 20 mM EDTA) without ╬▓-mercaptoethanol and homogenized using disposable pestles for microtubes. Samples were centrifuged at room temperature for 2 min at 10,000 ├Śg, then the liquid phase was transferred to a new microtube. Ethanol (150 ╬╝l) was added, the sample mixed, then quickly transferred to a column (FavorPrep Plasmid DNA extraction column, FAVORGEN, Ping Tun, Taiwan), centrifuged as above, but for 1 min, then the flow-through was discarded. The column was washed twice by centrifugation in 3 M Na acetate (450 ╬╝l), then in 70% ethanol (320 ╬╝l); the flow-through was again discarded. RNA was then eluted by centrifugation in sterile water (50 ╬╝l).
The nucleotide sequences of the coat protein of allexiviruses (garlic viruses A, B, C, D, and E) were downloaded from GenBank and aligned using MUSCLE version 3.8.31 (Edgar, 2004). Pairwise nucleotide identity of each allexivirus was calculated using MEGA version 11 (Tamura et al., 2021) and R version 4.2.1 (R Core Team, 2022).
The target cDNAs for the three viruses, with the actin gene as an internal control were amplified by one-step multiplex RT-PCR using PrimeScript One Step RT-PCR Kit version 2 (Dye plus) (TAKARA, Tokyo, Japan). The virus-specific forward primers were labeled with FITC and reverse primers were labeled with biotin (Table 1). For PCR, the thermal cycling conditions were 94┬░C for 2 min; 35 cycles of 94┬░C 30 s, 56┬░C 30 s, and 72┬░C 30 s; and 72┬░C for 10 min. For on-site detection of garlic viruses, a mobile PCR device (miniPCR mini16, miniPCR Bio, Cambridge, MA, USA) was used.
RT-PCR products were separated by 2% agarose gel electrophoresis (0.5├Ś TAE). In the dark (or in a light-resistant box), the gel was illuminated with LED blue light during electrophoresis. The cDNA band for each target virus was identified in a real-time manner. For on-site detection, an electrophoresis apparatus with a pre-installed blue light was used (blueGel electrophoresis system; miniPCR Bio).
The array assay was done essentially as we described previously (Furuta et al., 2017; Shimura et al., 2015; Sugiyama et al., 2008). The array membranes to detect multiple garlic viruses were designed and prepared by Hokusan Co. Ltd. (Hokkaido, Japan).
The cDNAs of the three garlic viruses were separately amplified in a uniplex one-step RT-PCR mixture containing primer pairs (0.5 ╬╝M each). The Universal Lateral Flow Assay Kit (Cytodiagnostics Inc., Burlington, ON, Canada) was used for the LFA. RT-PCR products were applied to the dipstick tips according to the manufacturerŌĆÖs instruction. After 15 min at room temperature, the samples were assessed for the presence or absence of the color line between the test and control.
Grinding of garlic tissues can be easily performed using disposable pestles and microtubes so that it can be performed even on-site. Subsequent RNA extraction can be also easily performed by a portable centrifuge without the use of a kit, but with an inexpensive single column, which is commercially available on a stand-alone base; for example, RNA extraction from 10 individual samples can be completed in as little as 15 min.
For developing a simple, inexpensive on-site detection system for the three garlic viruses, we first investigated the conditions for multiplex RT-PCR to enable simultaneous detection of the three viruses. Particularly difficult is the reliable detection of allexiviruses. There are currently seven species of allexiviruses, and the homology between the isolates is relatively low compared to the other viruses even though they are classified as the same species; the sequence variabilities of isolates belonging to garlic virus A (GarV-A) and GarV-C were found to be especially high (Fig. 1). Since our goal is to produce virus-free garlic in Hokkaido, we first sequenced the allexiviruses that infect garlic in Hokkaido (and some other regions in Japan), considering sequence variabilities specific to the Japanese (even Hokkaido) isolates. Based on this information, we created primers specific to each of the Hokkaido isolates and used them in the multiplex reaction system; we did not use any universal primer for allexiviruses. The multiplex PCR thus included 14 primers: eight for allexiviruses, two each for LYSV, OYDV, and the actin gene (Table 1). Here, to prevent the formation of primer dimers, the 3ŌĆÖ end sequences were manually adjusted to determine the optimal sequence combination.
Some animal viruses have been simultaneously detected by agarose gel electrophoresis after a multiplex RT-PCR using primers labeled with fluorescent dyes such as FITC (Shigemoto et al., 2011). However, for some reason, this method has rarely been applied to detecting plant viruses, perhaps due to the complexity of electrophoresis. Recently, however, convenient electrophoresis equipment has become available at low cost. For example, the blueGel electrophoresis system (miniPCR Bio), which we used here, makes electrophoretic detection much less complicated (Gonz├Īlez-Gonz├Īlez et al., 2020). Viral bands can be detected in real time within minutes by illuminating a blue light during electrophoresis. Thus, the time and labor required for electrophoresis and gel staining can be greatly reduced. Fig. 2 shows the flow of the FDA operation. We used a mobile PCR device that can be operated with a smartphone. A small electrophoresis device equipped with a blue light can also be handmade. Supplementary Fig. 1 shows a photograph of each step in the actual on-site detection of viruses using our system in the rear space of a van.
If there is a sample with a +/ŌłÆ result by the FDA method, the same PCR reaction mixture can also be used directly in the second step (array assay) to confirm the results of the first step by hybridization of nucleic acids. The array assay is simple to do because all reactions are performed in microtubes, and the results can be observed by eye (Supplementary Fig. 2). We have been using this array assay to detect several plant pathogens without any problems.
Because the FDA method uses RT-PCR, the detection sensitivity of the virus is considered equivalent to that of the conventional RT-PCR. To be sure, RT-PCR was performed using non-FITC-labeled primers, and the gel was stained with ethidium bromide (EB). The result is shown in Supplementary Fig. 3 where the FDA result using FITC-labeled primers is also placed next to the EB-stained gel for comparison. We judged that there was almost no difference in detection sensitivity between the two.
The specificity of the FDA-array can be confirmed by proceeding to the array assay following the FDA, which identifies the sequence of each virus by nucleic acid hybridization. To further demonstrate the high specificity of the FDA-array, the PCR products of the FDA were excised from the gel and sequenced. As a result, as shown in Supplementary Table 1, each virus sequence was specifically identified according to the size of the PCR product.
Fig. 3 shows a farmer using our detection system for garlic viruses in a field in Hokkaido, Japan, at the back of a van. Each virus band was distinguished without any problem (Fig. 3A). In Fig. 3B, the same PCR reaction mixture used in Fig. 3A was used for the array assay, and the final diagnosis is shown with the array results (Fig. 3B and C). As a result, the judgment for most of the samples was found to be identical between the two methods, except for three bands that were judged to be positive using the FDA method but were nonspecific in the array. It is important for garlic farmers to determine how much virus is present in their fields; however, 100% accuracy is not always necessary. Thus, we believe that the FDA method alone is sufficient for on-site detection of garlic viruses.
The FDA-array we here developed is especially designed for easy and rapid on-site detection of viruses infecting garlic in Hokkaido, Japan. However, we here conducted an additional experiment to assess the versatility of this current version. Garlic samples from various regions in Japan were tested using this current version of the FDA-array (Supplementary Fig. 4). As a result, this FDA method showed better applicability than what we had expected. In the future, it may become necessary to modify the primer design to detect the virus which has some sequence variability.
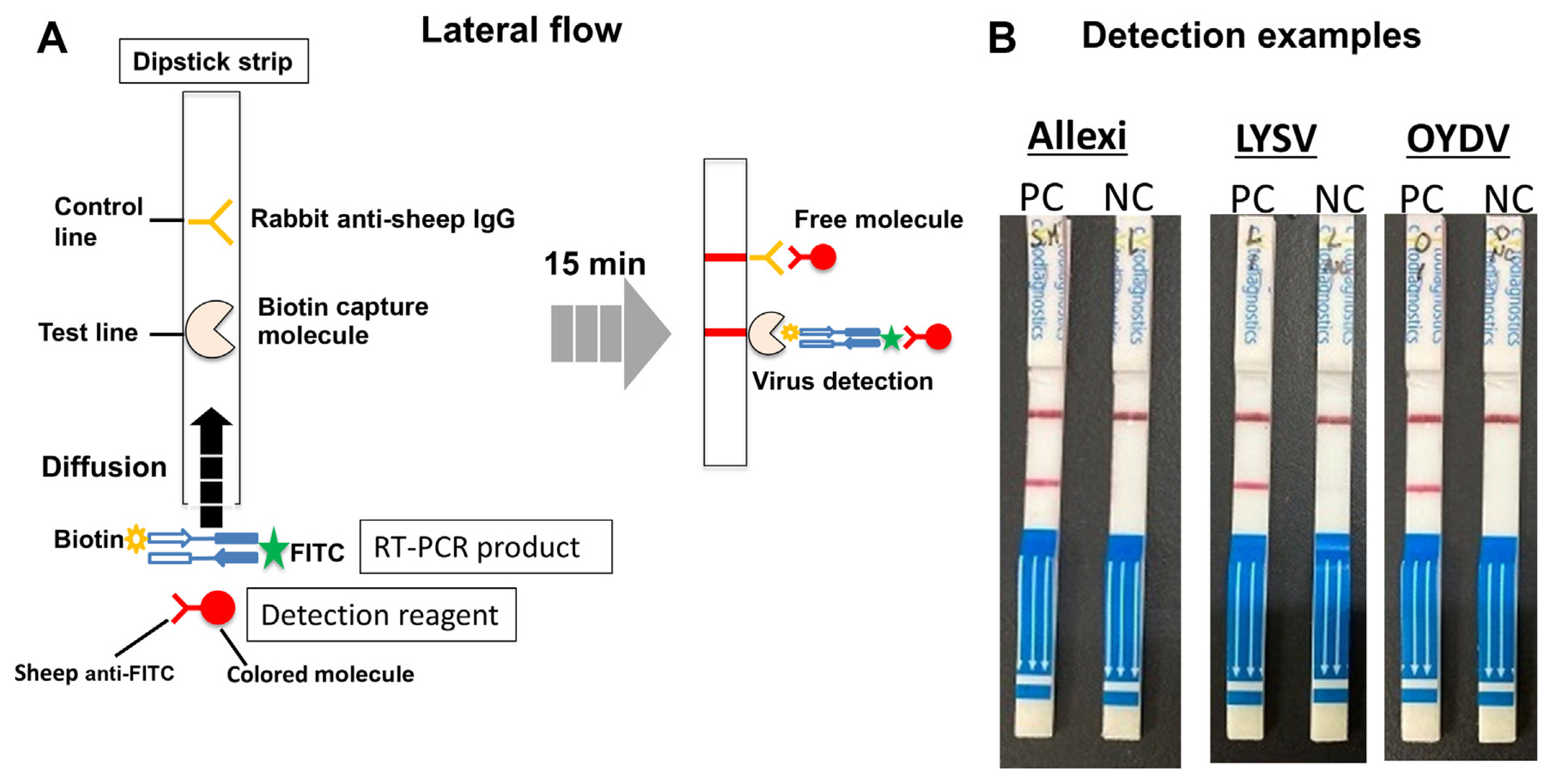
All reagents and kits required for the FDA method and the array assay can be supplied upon request by Hokusan Co. Ltd. or easily handmade. The only concern is to design primers for multiplexing properly. We considered that, at present, simple PCR and electrophoresis detection is the cheapest way for on-site detection of garlic viruses, but we are expecting to introduce an LFA to further simplify the detection. As shown in Fig. 4, the method to detect each of the three viruses using commercially available strips for LFA is simple, easy, and quick. However, since simultaneous diagnosis of at least three viruses is essential for garlic viruses, a prototype of a multiplex-LFA system is currently under development in our laboratory. Another attractive alternative to PCR is the use of the RPA, which can amplify nucleic acids at room temperature. However, this system is also very expensive at present, so we consider that it will take some time before it can be used for on-site detection.
Diagnosis of plant viruses can be divided into two main categories, depending on its purpose. One is diagnosis at the laboratory, research institute, or university level, where the emphasis is on accurate, highly sensitive detection; the use of expensive machinery, reagents and kits is not considered as a major problem. The second is on-site diagnosis at the farmer or agricultural organization level, where the emphasis is on low cost and simplicity of operation as long as a certain level of accuracy and sensitivity are guaranteed. On-site diagnosis often involves a large number of samples, and the first priority is to determine whether the suspected virus(es) is widespread in the field. At the laboratory level, RT-PCR is currently the most common method of diagnosis. For the on-site diagnosis, the RIPA method has been used in the past. However, its accuracy could be problematic because the antibody strips are not stable for long-term storage, and the success or failure of detection depended on the virus concentration in the plants. The technology for LFA has become recently popular for detecting coronaviruses and also is being used to detect even plant viruses. There are two types of LFA; one detects viral particles using an antigen-antibody reaction, and the other detects viral nucleic acids after amplification, which is more sensitive than the antigen-antibody reaction. Recently, RPA, which can amplify viral nucleic acid sequences at room temperature, was developed and combined with LFA as the RPA-LFA method for on-site detection of some plant viruses (Bhat et al., 2022). However, this method still needs to be improved. The available commercial kits are quite expensive for on-site diagnosis of plant viruses. In addition, diagnosis of plant viruses often requires simultaneous detection of multiple viruses, and the greater the number of primers, the more likely a nonspecific reaction will occur. In this regard, the RPA method, which is operated at room temperature, is thought to be more likely to produce nonspecific reactions than the conventional RT-PCR.
Therefore, we have now developed the FDA-array method in anticipation of a transitional period until RPA-LFA is available at a lower cost. The FDA is a step in the amplification of viral nucleic acid sequences by RT-PCR, but there is little problem in using it for on-site diagnosis with an inexpensive mobile PCR device. In addition, electrophoresis takes only several minutes, enabling a system that can detect viral bands in real time. The electrophoresis system can also be purchased inexpensively (or handmade). The only weakness compared to RT-RPA is that it requires about 2 h or less for RT-PCR. However, because the purpose of field diagnosis is often to determine the extent of spread of a suspected virus, this amount of time may not be of much concern.
Recently, we have seen many reports on the LAMP method, which claims to detect viruses with high sensitivity. However, there is still no report of the LAMP method being used to detect garlic viruses. Although the method is suitable for a laboratory diagnosis, it may have some drawbacks for field-level diagnosis for plant viruses. For example, (1) kits are expensive, (2) multiplexing is difficult to design although multiplex LAMP has been developed for some animal viruses, and (3) the method is very sensitive to contamination, which is a critical matter for on-site multiplex detection.
We expect that this FDA-array as a virus diagnostic system will be especially useful for field diagnosis of garlic viruses. We are now testing FDA-array for the multiplex detection of plant viruses other than garlic viruses. In the near future, we should be able to replace the electrophoresis step with multiplex-LFA. We thus propose a diagnostic system where users can choose the method depending on the purpose of viral detection (Supplementary Fig. 5).
Acknowledgments
We thank Dr. Hangil Kim for reviewing this manuscript. We also thank Drs. Minoru Takeshita and Masamichi Isogai for providing us garlic samples.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig.┬Ā1
Heatmap for the sequence variability matrix within the allexivirus species. The heatmap for each allexivirus (GarV-A, GarV-B, GarV-C, GarV-D, and GarV-E) is shown. Each matrix represents the nucleotide pairwise genetic distance computed under the Tamura-Nei model (GarV-A, n = 89; GarV-B, n = 45; GarV-C, n = 24; GarV-D, n = 74; GarV-E, n = 10).

Fig.┬Ā2
Flowchart of RNA extraction and FDA for garlic viruses. Total RNA was extracted by grinding garlic tissues in the extraction buffer using a disposal pestle in a microtube, and the tissue debris was removed by centrifugation using a portable table centrifuge. RNA from garlic samples is used to amplify viral cDNAs by RT-PCR with FITC- and biotin-labeled virus-specific primer pairs. FITC-labeled RT-PCR products are then separated using agarose gel electrophoresis. The PCR bands can be identified in real time without staining using a gel electrophoresis system equipped with an LED blue light illuminator. FDA, FITC-detection assay; RT-PCR, reverse transcription polymerase chain reaction; PCR, polymerase chain reaction.
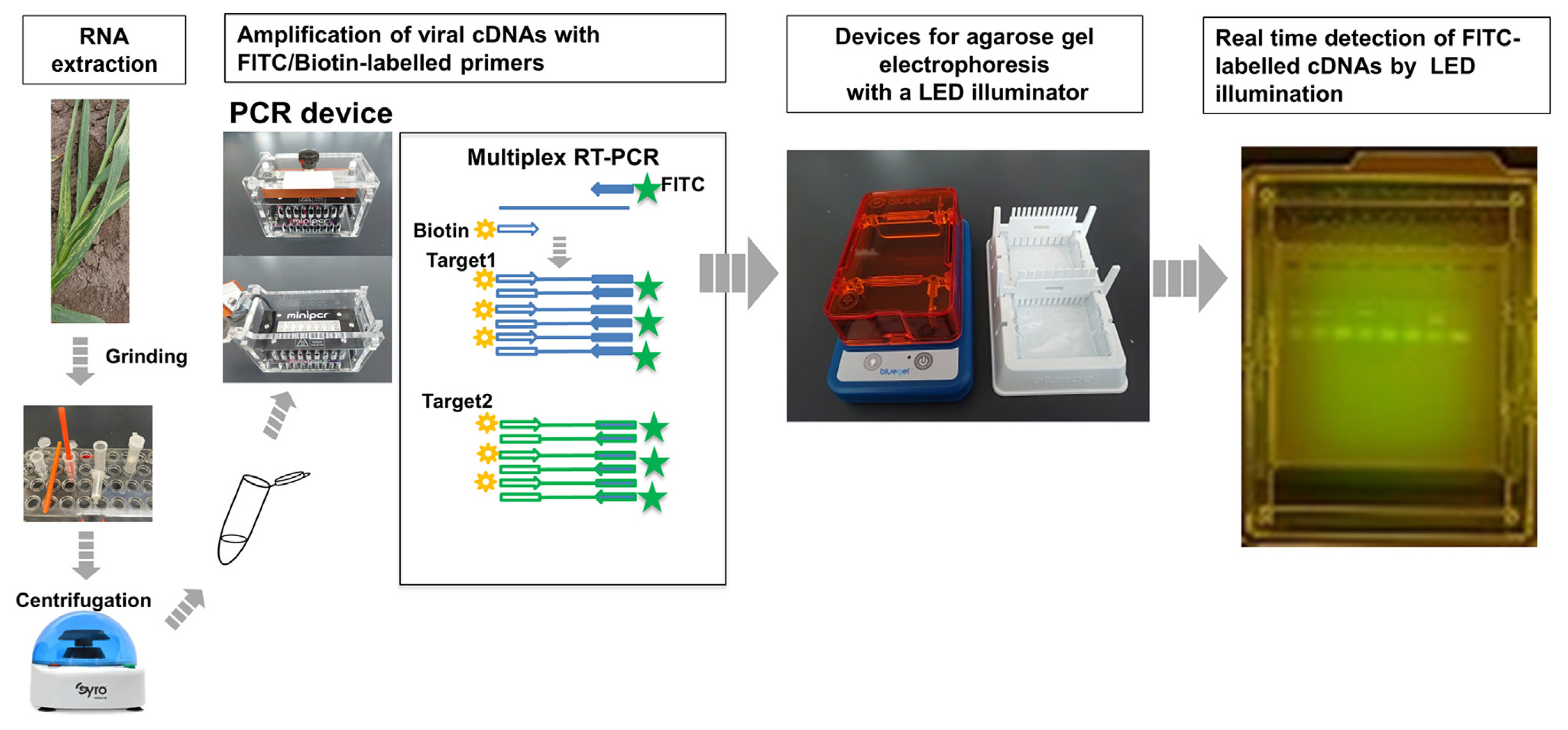
Fig.┬Ā3
On-site detection of garlic viruses in the field of Hokkaido using the FDA-array method. (A) Example of the results of the FDA part to detect three target garlic viruses, allexiviruses, LYSV, and OYDV. The judgment for the presence (+) or absence (ŌłÆ) of the PCR band specific for each virus is shown below the gel. (B) Example of the results for the array (signals on the membranes). The RT-PCR products from the FDA were directly used in the next detection step, the array assay. Samples included in each lane are as follows. Lane M, size marker, mixture of the PCR products of the three garlic viruses amplified with FITC-labeled and biotin-labeled primer pairs. FDA, FITC-detection assay; Allexi, allexiviruses; LYSV, leek yellow stripe virus; OYDV, onion yellow dwarf virus; PCR, polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction. Lanes 1-16, samples from the garlic fields of Tokachi area, Hokkaido; NC, negative control (distilled water); lanes P1-6, garlic bulbs from the market; P1, Chinese; P2, Japanese; P3-5, Chinese; P6, Spanish. (C) Final diagnosis on presence (+) or absence (ŌłÆ) of each virus.

Fig.┬Ā4
Flowchart for garlic virus detection using the lateral flow method. (A) Principle of virus detection. The Universal Lateral Flow Assay Kit from Cytodiagnostics Inc. (Burlington, ON, Canada) was used. The dipstick strip has a test line with a fixed biotin capture molecule and a control line with a fixed rabbit anti-sheep IgG. Biotin- and FITC-labeled RT-PCR products and anti-FITC sheep IgG with colored molecules migrate through the strips. RT-PCR products captured on the test line and the unreacted FITC primers captured on the control line are bound by anti-FITC IgG so that the lines develop color. (B) Examples of positive results for the detection of three viruses. Viral cDNA was amplified by a uniplex RT-PCR with FITC- and biotin-labeled primer pairs to detect each virus. RT-PCR, reverse transcription polymerase chain reaction; PC, virus-infected leaf; NC, water; Allexi, allexiviruses; LYSV, leek yellow stripe virus; OYDV, onion yellow dwarf virus.
Table┬Ā1
Primers used in this study
| Virus | Primer | Direction | Primer sequence | Product size (bp) |
|---|---|---|---|---|
| Allexivirusa | Allexi-array-Fa | Forward | ACTGACGCGCCTTGCGGCAT | 710-730 |
| Allexi-array-Fb | Forward | ACGGATGCCCCTTGCGGCAT | ||
| Allexi-array-Fc | Forward | ACGGATGCGCCGTGTGGAAT | ||
| Allexi-array-Fd | Forward | ACGAAAGCACCTTGCGGAAT | ||
| Allexi-array-Fe | Forward | ACCAATGCACCCTGTGGCAT | ||
| Allexi-array-Fx | Forward | ACTGACGCACCTTGTGGCAT | ||
| Al-cp3-750-1 | Reverse | CCCTTCAGCATATAGCTTAGC | ||
| Al-cp3-750-2 | Reverse | CCTTTCAGCATATAGCTTAGC | ||
| OYDV | OYDV-F | Forward | GTGATGCAGCTGAAGCATACA | 250 |
| OYDV-R | Reverse | GCTGTGTGTCTTTCCGTGTCCTCTTC | ||
| LYSY | LYSY-F | Forward | ACTCGTTCAAGGCAGTTTCT | 401 |
| LYSV-R | Reverse | ACCACGCTACACCAGTAAATAA | ||
| Actin (control) | Gar-Act-F | Forward | GAATTGTGAGCAACTGGGATGAC | 226 |
| Gar-Act-R | Reverse | GTGGTACGGCCACTGGCATA |
References
Bhat, A. I., Aman, R. and Mahfouz, M. 2022. Onsite detection of plant viruses using isothermal amplification assays. Plant Biotechnol. J. 20:1859-1873.




Chen, J., Zheng, H.-Y., Antoniw, J. F., Adams, M. J., Chen, J.-P. and Lin, L. 2004. Detection and classification of allexiviruses from garlic in China. Arch. Virol 149:435-445.



Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797.



Furuta, K., Nagashima, S., Inukai, T. and Masuta, C. 2017. Construction of a system for the strawberry nursery production towards elimination of latent infection of anthracnose fungi by a combination of PCR and microtube-hybridization (PCR-MTH). Plant Pathol. J. 33:80-86.


Gonz├Īlez-Gonz├Īlez, E., Trujillo-de Santiago, G., Lara-Mayorga, I. M., Mart├Łnez-Chapa, S. O. and Alvarez, M. M. 2020. Portable and accurate diagnostics for COVID-19: combined use of the miniPCR thermocycler and a well-plate reader for SARS-CoV-2 virus detection. PLoS ONE 15:e0237418.



Hu, X.-X., Lei, Y., Wang, P., Tang, L.-F., He, C.-Z., Song, Y., Xiong, X.-Y. and Nie, X.-Z. 2015. Development of a multiplex reverse transcription-PCR assay for simultaneous detection of garlic viruses. J. Integr. Agric. 14:900-908.

Jayasinghe, W. H., Kim, H., Sasaki, J. and Masuta, C. 2021. Aphid transmissibility of onion yellow dwarf virus isolates with an N-terminal truncated HC-Pro is aided by leek yellow stripe virus. J. Gen. Plant Pathol. 87:178-183.


Kawakubo, S., Kim, H., Takeshita, M. and Masuta, C. 2023. Host-specific adaptation drove the coevolution of leek yellow stripe virus and Allium plants. Microbiol. Spectr 11:e0234023.




Kim, D.-H., Jeong, R.-D., Choi, S., Ju, H.-J. and Yoon, J.-Y. 2022a. Application of rapid and reliable detection of cymbidium mosaic virus by reverse transcription recombinase polymerase amplification combined with lateral flow immunoassay. Plant Pathol. J. 38:665-672.




Kim, H., Kawakubo, S., Takahashi, H. and Masuta, C. 2023. Two mutually exclusive evolutionary scenarios for allexiviruses that overcome host RNA silencing and autophagy by regulating viral CRP expression. PLoS Pathog. 19:e1011457.



Kim, N.-K., Lee, H.-J., Kim, S.-M. and Jeong, R.-D. 2022b. Rapid and visual detection of barley yellow dwarf virus by reverse transcription recombinase polymerase amplification with lateral flow strips. Plant Pathol. J. 38:159-166.




Majumder, S. and Baranwal, V. K. 2014. Simultaneous detection of four garlic viruses by multiplex reverse transcription PCR and their distribution in Indian garlic accessions. J. Virol. Methods 202:34-38.


Nam, M., Lee, Y.-H., Park, C. Y., Lee, M.-A., Bae, Y.-S., Lim, S., Lee, J. H., Moon, J. S. and Lee, S.-H. 2015. Development of multiplex RT-PCR for simultaneous detection of garlic viruses and the incidence of garlic viral disease in garlic genetic resources. Plant Pathol. J. 31:90-96.



Park, K.-S., Bae, Y.-J., Jung, E.-J. and Kang, S.-J. 2005. RT-PCR-based detection of six garlic viruses and their phylogenetic relationships. J. Microbiol. Biotechnol. 15:1110-1114.
R Core Team 2022 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. URL https://www.Rproject.org/ [1 November 2023].
Sasaki, J., Kawakubo, S., Kim, H., Kim, O.-K., Yamashita, K., Shimura, H. and Masuta, C. 2022. Leek yellow stripe virus can adjust for host adaptation by trimming the N-terminal domain to allow the P1 protein to function as an RNA silencing suppressor. Plant Pathol. J. 38:383-394.




Shigemoto, N., Fukuda, S., Tanizawa, Y., Kuwayama, M., Ohara, S. and Seno, M. 2011. Detection of norovirus, sapovirus, and human astrovirus in fecal specimens using a multiplex reverse transcription-PCR with fluorescent dye-labeled primers. Microbiol. Immunol 55:369-372.


Shimura, H., Furuta, K. and Masuta, C. 2015. Detection of plant viruses in mixed infection by macroarray-assisted method. Methods Mol. Biol. 1236:1-11.

Sugiyama, S., Masuta, C., Sekiguchi, H., Uehara, T., Shimura, H. and Maruta, Y. 2008. A simple, sensitive, specific detection of mixed infection of multiple plant viruses using macroarray and microtube hybridization. J. Virol. Methods 153:241-244.


Tamura, K., Stecher, G. and Kumar, S. 2021. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 38:3022-3027.




Yaffe, H., Buxdorf, K., Shapira, I., Ein-Gedi, S., Zvi, MM-B, Fridman, E., Moshelion, M. and Levy, M. 2012. LogSpin: a simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes 5:45.




Yoshida, N., Shimura, H. and Masuta, C. 2018. Allexiviruses may have acquired inserted sequences between the CP and CRP genes to change the translation reinitiation strategy of CRP. Arch. Virol 163:1419-1427.



- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 809 View
- 108 Download
- ORCID iDs
-
Chikara Masuta

https://orcid.org/0000-0003-3129-9167 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



