 |
 |
| Plant Pathol J > Volume 32(3); 2016 > Article |
Abstract
Together with the Fusarium graminearum species complex, F. culmorum is a major member of the causal agents of Fusarium head blight on cereals such as wheat, barley and corn. It causes significant yield and quality losses and results in the contamination of grain with mycotoxins that are harmful to humans and animals. In Korea, F. culmorum is listed as a quarantine fungal species since it has yet to be found in the country. In this paper, we report that two isolates (J1 and J2) of F. culmorum were collected from the air at a rice paddy field in Korea. Species identification was confirmed by phylogenetic analysis using multi-locus sequence data derived from five genes encoding translation elongation factor, histone H3, phosphate permease, a reductase, and an ammonia ligase and by morphological comparison with reference strains. Both diagnostic PCR and chemical analysis confirmed that these F. culmorum isolates had the capacity to produce nivalenol, the trichothecene mycotoxin, in rice substrate. In addition, both isolates were pathogenic on wheat heads and corn stalks. This is the first report on the occurrence of F. culmorum in Korea.
Fusarium culmorum is a soil-borne ascomycetous fungus distributed in cooler temperate zones, including northern, central, and Western Europe, Canada, Western Asia, North Africa, and Australia (Demeke et al., 2005; Iwama et al., 2007; Obanor et al., 2010; Parry et al., 1995; Stepień et al., 2008; Yli-Mattila, 2010). It is a causal agent of serious diseases, such as crown rot, ear rot, foot rot, and head blight in cereals (e.g., wheat, barley, and corn) (Obanor et al., 2010; Scherm et al., 2013; Wagacha and Muthomi, 2007). It is also known to be one of the two main pathogens causing Fusarium head blight (FHB) on wheat and barley, along with the F. graminearum species complex (Fg complex) (Goswami and Kistler, 2004; Hogg et al., 2010; Miedaner et al., 2008; Scherm et al., 2013; Wagacha and Muthomi, 2007). Recently, the composition of both Fusarium species has changed in Europe and Australia, probably due to climate change (Jennings et al., 2004; Miedaner et al., 2008; Obanor et al., 2010; Scherm et al., 2013; Waalwijk et al., 2003; West et al., 2012). F. culmorum can survive in the soil and infect the roots of cereal plants; its asexual spores (conidia) can also infect plants through the florets. In addition to causing plant diseases, F. culmorum produces several mycotoxins on infected cereals, such as trichothecenes and zearalenone, which are harmful to humans and animals; the trichothecenes can be divided into two chemotypes (deoxynivalenol, DON; nivalenol, NIV), as in the Fg complex (Bakan et al., 2001; Demeke et al., 2005; Nielsen et al., 2012; Pasquali et al., 2010; Scherm et al., 2013). Unlike the Fg complex, F. culmorum is not able to undergo sexual reproduction for the production of sexual progeny (Kerényi et al., 2004; Mishra et al., 2003; Obanor et al., 2010), and is not recognized as a species complex since its global populations lack a strong lineage structure despite their geographic separation (Obanor et al., 2010). In Korea, F. culmorum is listed as a fungal quarantine pest since it has not been found in the country (Animal, Plant and Fisheries Quarantine and Inspection Agency, 2013). In this study, we identified two isolates of putative F. culmorum, which were isolated from the air at a rice paddy field in Korea, by both morphological and molecular comparisons to reference strains of F. culmorum, and assessed their potential pathogenicity and mycotoxin production. This is the first report of the occurrence of F. culmorum in Korea.
Airborne Fusarium species were collected as previously described (Jung et al., 2013). Ninety-millimeter Petri dishes of pentachloronitrobenzene (PCNB) agar medium (1.5% peptone, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.075% PCNB, 2% agar) supplemented with kanamycin (75 μg/ml) were exposed at a farm road next to a rice paddy field located in Gigokri, Dogo-myeon, Asan, Chungnam Province, Korea on April 4, 2015, which was not close to corn-growing fields and has not been used for wheat cultivation. Three PCNB plates were left open on a 1-m-high stand for 30 minutes at three different locations approximately 100 m apart from each other. The plates were then incubated at 25ºC for three days, and the fungal colonies were sub-cultured onto fresh potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA). Each Fusarium isolate was purified through single-spore isolation. Reference strains of Fusarium were obtained from the Centraalbureau voor Schimmelcultures (Utrecht, the Netherlands). Several species belonging to the Fg complex such as F. graminearum, F. aisaticum, F. botthii, F. vorosii, and F. fujikuroi used in this study were previously described (Kim et al., 2015), or obtained from our fungal collection. Fungal strains from 25% glycerol stock cultures stored at −80ºC were maintained on PDA. To analyze colony morphology and hyphal growth, the Fusarium strains were grown in either PDA or complete medium (CM; Leslie and Summerell, 2006) at 25ºC. To observe conidial morphology, agar blocks of the fungal hyphae were inoculated on carnation leaf agar (CLA) medium prepared as described previously, and incubated for two weeks under black light (Leslie and Summerell, 2006). For genomic DNA extraction, the Fusarium species were grown in 50 ml of CM broth at 25ºC for four days with shaking (150 rpm). For trichothecene production, agar blocks from CM culture of the F. culmorum isolates were inoculated into 40 g of autoclaved rice in a 500-ml Erlenmeyer flask and incubated for three weeks. Fungal genomic DNA was extracted by a quick method as described previously (Chi et al., 2009).
For identification of the species of F. culmorum isolates and their phylogenetic relationships with other F. culmorum strains worldwide, partial nucleotide sequences of the five genes frequently used as DNA markers (those for translation elongation factor 1-alpha [TEF1, ~680 bp], histone H3 [H3, ~450 bp], phosphate permease [PHO, ~850 bp], reductase-like gene [RED, ~820 bp], and ammonia ligase [URA, ~570 bp]) were amplified from the collected Fusarium isolates as previously described (O’Donnell et al., 2000). For trichothecene chemotype determination, the TRI7 and TRI13 genes were amplified as previously described (Lee et al., 2001). For mating-type determination, two primer sets, which were derived from F. graminearum and specific to the MAT1-1 or MAT1-2 locus, were used. All of the PCR primers (Table 1) (Chandler et al., 2003; O’Donnell et al., 2000) used in this study were synthesized by Cosmo Genetech (Seoul, Korea). Each reaction tube contained 50 ng of template DNA, 1 × PCR buffer, dNTPs at 0.2 mM each, primers at 10 μM, and 1.25 U ExTaq polymerase (Takara Biomedicals, Shiga, Japan) in 50-μl reaction volumes. PCR products of the DNA marker genes for phylogenetic analysis were sequenced directly after purification with a PCR clean-up system (Promega, Madison, WI, USA). For phylogenetic analysis, the nucleotide sequences for each marker gene from the Korean isolates were combined and aligned with those of the reference strains retrieved from GenBank (O’Donnell et al., 2000) using ClustalW (Thompson et al., 1994). A total of 2,751 nucleotides from all five genes, or 2,212 nucleotides from three genes (PHO, RED, and TEF1), were used in maximum likelihood (ML) analyses using MEGA ver. 4.02 (Center of Evolutionary Functional Genomics Biodesign Institute, Arizona State University, Tempe, AZ, USA). The robustness of the ML trees (MLTs) was determined using the full heuristic search option for 1,000 bootstrap replications. The DNA sequences of the genes newly obtained from Korean F. culmorum isolates and two reference strains were deposited in GenBank (http://www.ncbi.nlm.nih.gov/) under the accession numbers listed in Table 2.
Quantitative determination of trichothecenes produced in the rice substrates was achieved by LC-MS/MS analysis. A homogenized rice sample (1 g) was extracted with 4 ml of acetonitrile:water (50:50) containing 0.1% formic acid and 1 g of sodium chloride. After filtering through a spin-X micro-centrifuge 0.2 μm filter (Sigma-Aldrich, Seoul, Korea), the supernatant was injected into the LC-MS/MS system consisting of an LC-10ADvp (Shimadzu, Kyoto, Japan) pump system with an API 2000 triple quadrupole mass spectrometer (AB SCIEX, Foster City, CA, USA) equipped with an ESI source. The Agilent ZORBAX ODS C18 column (4.6 × 25 cm, 5 μm particle size) was used for this work. The identification of precursor and product ions in positive mode was performed in selected reaction monitoring mode.
To test pathogenicity on wheat heads, a central spikelet of wheat was inoculated during mid-anthesis with a suspension of the fungal conidia at 105/ml, obtained from a three-day-old CMC culture as described previously (Han et al., 2007; Lee et al., 2009). To test pathogenicity of stalk rot on corn, plants grown in pots for two months were inoculated by injecting 10 μl of the suspension of fungal conidia (at 105/ml) using a micropipette tip at the second internode. Two weeks after inoculation, the corn stalks were cut longitudinally to identify stalk discoloration.
Two Korean isolates of F. culmorum (designated J1 and J2) were morphologically identified based on cultures grown on PDA and CLA. The J1 and J2 isolates exhibited colony features on PDA similar to those from the reference strains of F. culmorum (NRRL11427 and NRRL14983) and F. graminearum (PH-1 and Z3643): fast-growing, whitish, aerial hyphae with red pigmentation on the reverse plate on PDA and abundant macroconidia but no microconidia. However, the macroconidia of J1 and J2 formed on CLA showed morphological features typical of F. culmorum: slightly curved, rather short, thick-walled three- to four-septate, mostly measuring 32-35 × 4-5 μm, rounded and blunt-shaped apical cells lacking distinctive foot-shaped basal cells. The lengths and widths of macroconidia in J1 and J2 were highly similar to those from NRRL11427 and NRRL14983, but they were clearly shorter and larger, respectively, than those from Z3643 and PH-1 (Fig. 1). In addition, thick-walled oval to globose chlamydospores, which is the other key character for identifying F. culmorum, were formed in both hyphae and macroconidia on CLA (data not shown). Based on the culture morphology, F. culmorum has been frequently confused with F. sambucinum and F. cerealis (synonym F. crookwellense), but the relatively high growth rate and the notched basal cell shape of conidia in J1 and J2 clearly rule out this possibility (Leslie and Summerell, 2006).
The multilocus sequence (MLS) dataset from the five combined genes included 16 taxa and comprised 2,751 nucleotides. The MLT showed a topology that clearly separated F. culmorum and its closely related species, such as F. cerealis, F. pseudograminearum, and the members of the Fg complex, into four well-supported species clades (Fig. 2). In particular, the J1 and J2 isolates grouped with the F. culmorum reference strains (NRRL3288, NRRL14983, NRRL25475, and NRRL11427) in a strongly supported clade (bootstrap [BS] 100%); all of the examined reference strains of F. cerealis, which is a species frequently confused with F. culmorum based on its morphology, were placed in the other strongly supported clade (BS 100%). In addition, this MLS phylogeny clearly differentiated all of the examined isolates of the Fg complex into their respective phylogenetic subclades within the complex clade, with strong support (BS 72-100%) (Fig. 2). To assess the evolutionary relationship between the Korean F. culmorum isolates and global F. culmorum populations, ML analysis was performed using three combined genes (PHO, RED, and TEF1) derived from the previously characterized F. culmorum isolates present worldwide (Obanor et al., 2010). This three-gene dataset included 42 F. culmorum isolates mainly from Australia, Russia, Germany, Turkey, Tunisia, and Syria, and comprised 2,212 nucleotides. The MLT showed four clearly separated Fusarium species clades (F. culmorum, F. cerealis, F. pseudograminearum, and F. graminearum) (Fig. 3). For both J1 and J2 isolates, there was significant support for their placement in a subclade within the F. culmorum clade (BS 60%), which consisted mainly of isolates from Australia. In addition to this subclade, two more subclades were found among the global F. culmorum populations, one with BS 94% and the other with BS 62%. However, their phylogenetic positions were not significantly correlated with their geographic origins, as previously suggested (Obanor et al., 2010). To determine the similarity of J1 and J2 with Asian F. culmorum isolates, an MLT was constructed using only a single gene (TEF1), since no additional sequence information was available from the Asian isolates (Fig. 4). The topology of the MLT constructed using the TEF1 sequence was similar to that using the combined genes, and revealed that the J1 and J2 isolates formed a distinct subclade along with a Japanese isolate (MAFF 236454) (with BS 63%) within the F. culmorum clade, being significantly separated from its closely related species (F. graminearum, F. cerealis, and F. sambucinum). This indicates that the Korean F. culmorum isolates may have an evolutionary relationship with this Japanese isolate, but the similarity to the other Asian isolates (from China) is unclear (Fig. 4).
For determination of the species, mating type, and trichothecene chemotype of the J1 and J2 isolates, we performed diagnostic PCR using the specific primer sets listed in Table 1 (Fig. 5). An F. culmorum-specific primer set, derived from the nucleotide sequences of the TRI101 gene (unpublished data), successfully amplified an 820-bp fragment from the J1 and J2 isolates, as well as two F. culmorum reference strains (NRRL14983 and NRRL11427), but not from the representative strains of three species within the Fg complex and those from F. cerealis, F. sambucinum, and the distantly related F. fujikuroi (Fig. 5). The PCR with the mating-type locus-specific primer sets amplified diagnostic fragments for MAT1-2, but not MAT1-1, from both J1 and J2, indicating that these isolates carried only the MAT1-2 locus; the two F. culmorum reference strains, NRRL14983 and NRRL11427, were identified as MAT1-1 and MAT1-2, respectively (Fig. 6). The primer sets derived from TRI13 and TRI7, responsible for the DON/NIV chemotypes, amplified specific fragments from J1 and J2, which were identical to those from the NIV-producing F. asiaticum SCKO4 strain, suggesting that both J1 and J2 have the potential to synthesize NIV (Fig. 7).
LC-MS/MS analysis clearly confirmed the ability of both J1 and J2 isolates to produce NIV on rice substrate, at 1,062.4 μg/g for J1 and 678.4 μg/g for J2; DON production was not detected. However, the other two F. culmorum reference strains produced no detectable levels of NIV on rice. By comparison to previous reports of relatively high NIV production by several Fusarium isolates in cereal cultures, the NIV levels produced by both F. culmorum J1 and J2 isolates are at least two to three-fold higher (Gang et al., 1998; Muthomi et al., 2000). It was reported that several European F. culmorum isolates produced NIV at relatively high levels, up to 220.6 μg/g on cracked corn (by a German isolate; Muthomi et al., 2000) or up to 381.0 μg/g on rye grain (by a Dutch isolate; Gang et al., 1998). In addition, a Korean isolate of F. graminearum from barley (now re-identifiable as F. asiaticum), which is a closely related species to F. culmorum, exhibited a similar level of NIV production (416 μg/g) on a rice substrate (Kim et al., 1993). It was suggested that the wide distribution of F. graminearum (probably F. asiaticum) NIV chemotypes capable of producing high levels of NIV in rice paddy fields in Korea (Kim et al., 1993), where barley is cultivated in double-cropping systems, supports a close association of NIV-producing F. asiaticum isolates with rice-growing environments (Lee et al., 2009). In this respect, it is likely that the NIV-producing F. culmorum isolates presented in this study also have a close association with rice.
Pathogenicity tests revealed that both J1 and J2 were pathogenic, but showed different levels of virulence on several hosts. When J1 was inoculated onto wheat heads, typical head blight symptoms developed, while J2 caused no apparent symptoms (Fig. 8). However, both isolates caused typical stalk rot symptoms in corn; the necrotic regions caused by J1 seemed to be larger than those by J2 (Fig. 8).
The species identification of the J1 and J2 isolates were clearly confirmed by several approaches in this study: morphological comparison, phylogenetic analysis, and diagnostic PCR. This is thus the first report on the occurrence of F. culmorum in Korea. In addition, it should be noted that the occurrence of F. culmorum in the air has not been described previously. Although these Korean isolates were obtained from the air at a rice paddy field, not from a plant source, with a low frequency (found only once during a one-year survey), their capacities to cause disease symptoms and to produce NIV mycotoxin suggest the possibility of FHB on cereals caused by F. culmorum in Korea. Furthermore, their site of isolation (a rice paddy field), phylogenetic similarity to a Japanese F. culmorum isolate, and in vitro ability to produce NIV at relatively high levels may indicate that they have somehow been associated or adapted with rice in Korea, as the NIV-producing member (F. asiaticum) of the Fg complex is a major FHB pathogen (Lee et al., 2009). However, it is unclear where these airborne F. culmorum came from; it is likely that they originated from the fungal biomass associated with plant debris and/or in the soil since F. culmorum can survive in soil. To resolve this issue, it will be necessary to confirm the infection or contamination of rice and other cereals with F. culmorum in Korea, followed by a population study using more isolates from Korean F. culmorum populations.
Acknowledgments
This research was supported by grants from the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea, and from the Next-Generation Bio Green21 Program (No. PJ0111802015), the Rural Development Administration, Republic of Korea.
Fig. 1
Mycelial growth on complete medium (top and bottom views are shown in left and right panels, respectively, for each isolate) (A), macroconidial morphology (B-D), and size (E). J1 and J2, Korean Fusarium culmorum isolates; 11427 and 14983, NRRL strains of F. culmorum; PH-1 and Z43, F. graminearum PH-1 and Z3643 strains, respectively. Scale bars = 20 μm. Different letters above bars indicate significant differences.

Fig. 2
Maximum likelihood (ML) tree inferred from the combined five-locus dataset for Fusarium culmorum and its closely related species. The nucleotide sequences of F. pseudograminearum NRRL 28062 strain were used as the outgroup to root the phylogeny. ML bootstrap values are indicated above the internodes. The numbers of fungal isolates represent the NRRL number. The J1 and J2 isolates are in a gray-shaded box.
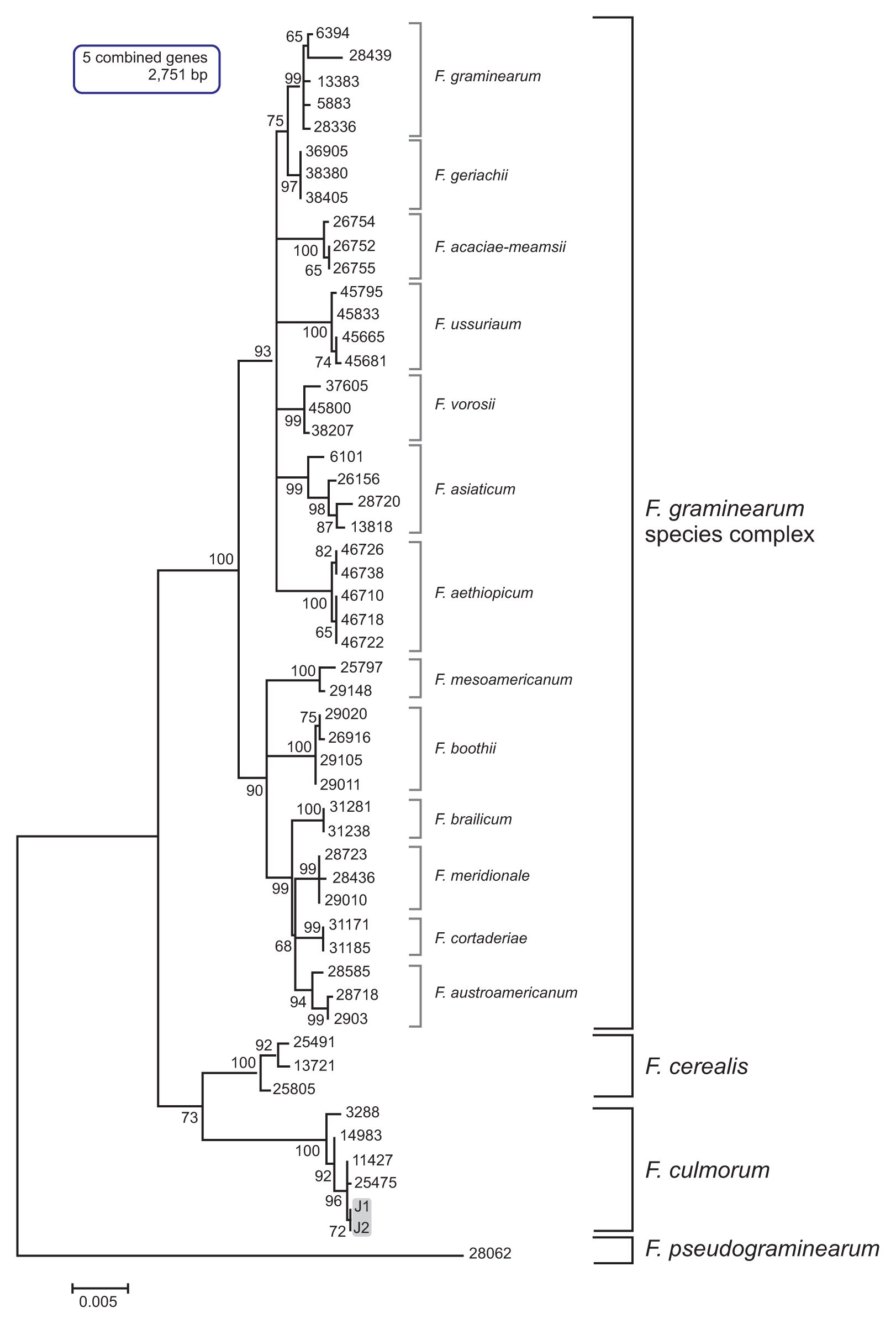
Fig. 3
Maximum likelihood (ML) tree inferred from the combined three-locus dataset for the global Fusarium culmorum populations. The nucleotide sequences of F. pseudograminearum strain were used as the outgroup to root the phylogeny. ML bootstrap values are indicated above internodes. The J1 and J2 isolates are in a gray-shaded box.
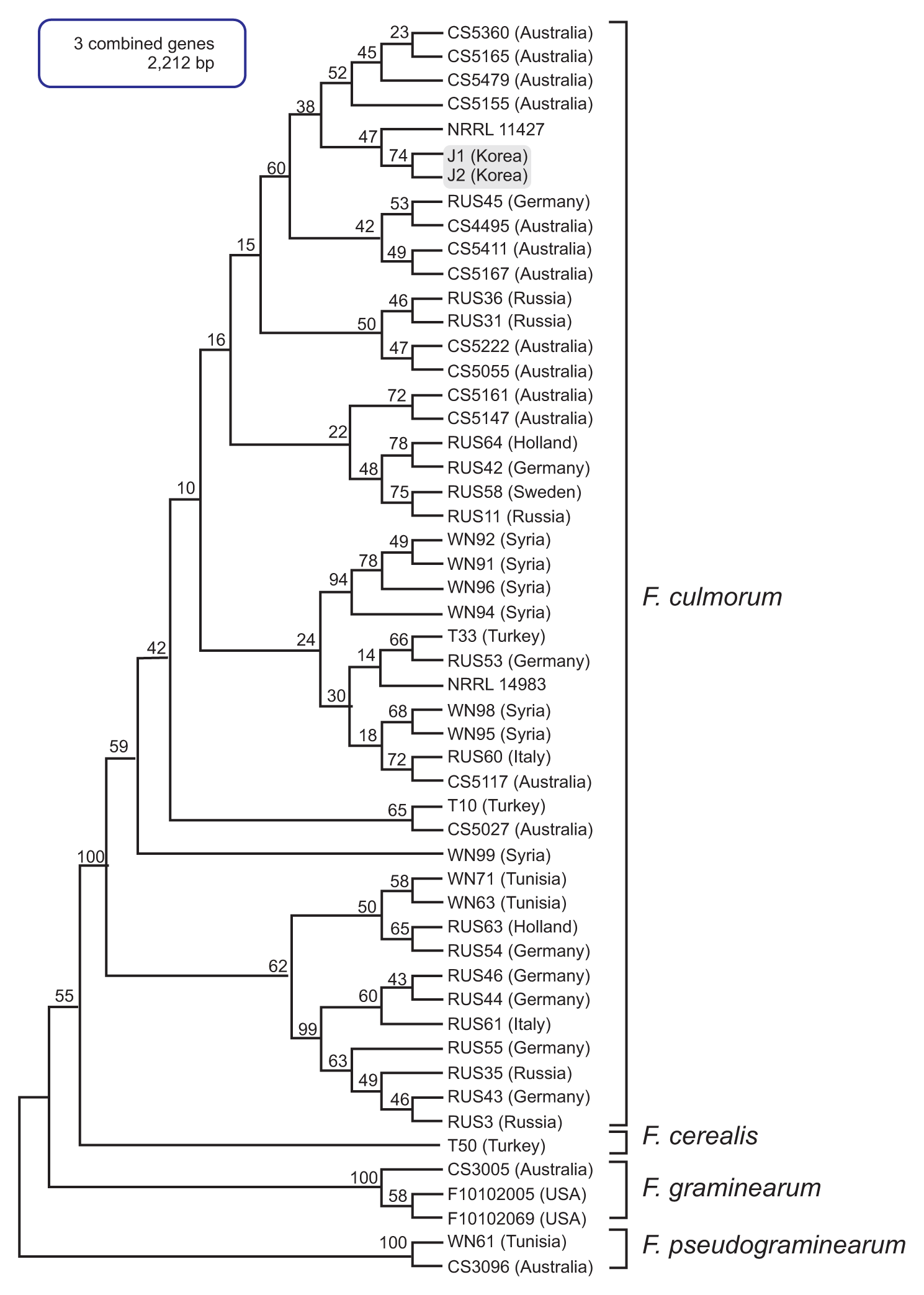
Fig. 4
Maximum likelihood (ML) tree inferred from the nucleotide sequences of TEF1 for the global Fusarium culmorum populations. The nucleotide sequences of F. sambucinum strain were used as the outgroup to root the phylogeny. ML bootstrap values are indicated above internodes. The J1 and J2 isolates are in a gray-shaded box.
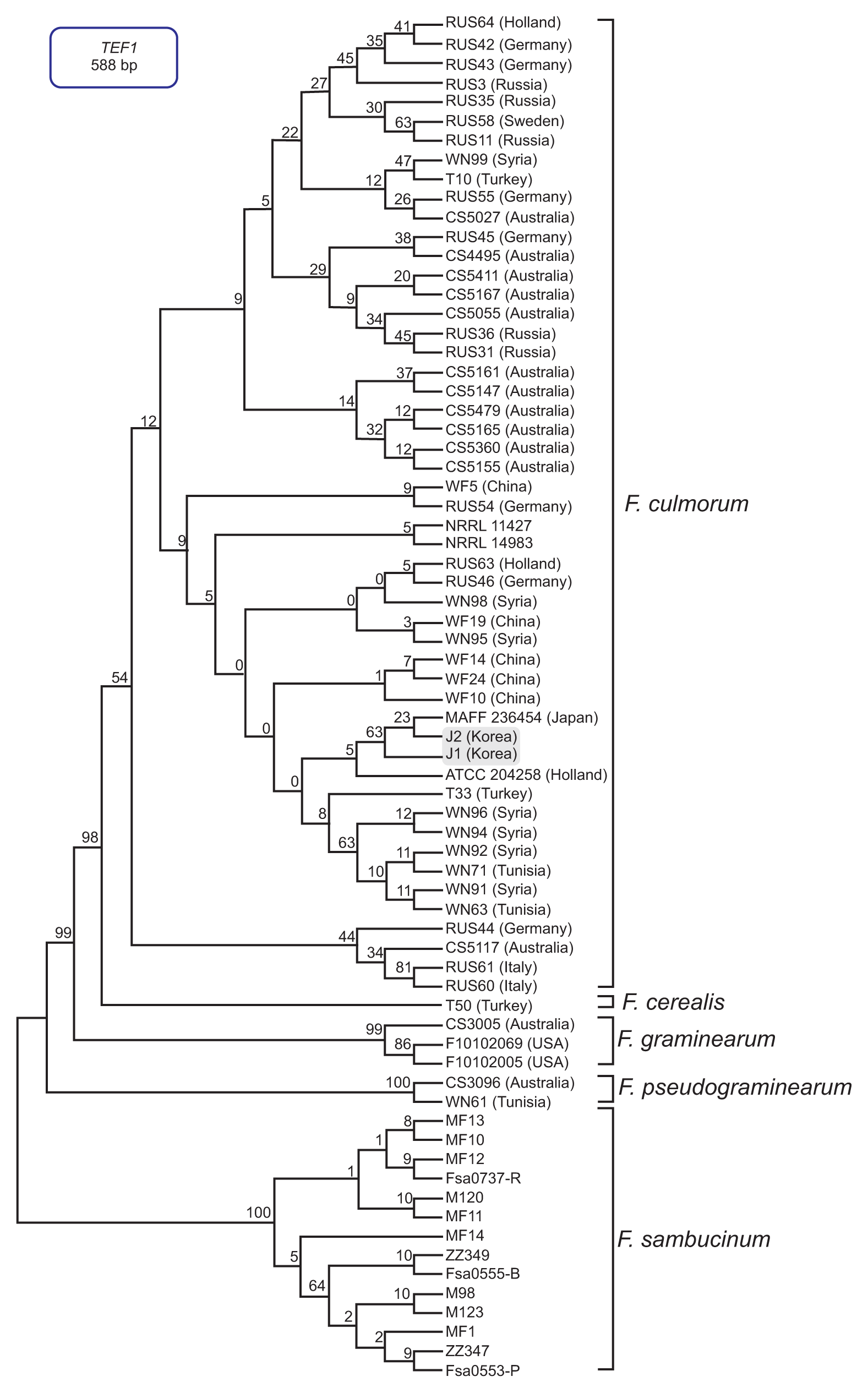
Fig. 5
PCR amplification with an Fusarium culmorum-specific primer pair (FculTri101 nF/p1 and FculTri101 nR/p1). Lanes 1, J1; 2, J2; 3, NRRL14983; 4, NRRL11427; 5, F. graminearum Z3643; 6, F. graminearum PH-1; 7, F. fujikuroi B14; 8, F. fujikuroi B20; 9, NRRL20481; 10, NRRL20482; 11, NRRL20483; 12, NRRL20484; 13, NRRL6574; 14, NRRL11451; 15, NRRL11453; 16, NRRL4833. Size markers (in kb) are indicated on the left of the gel.
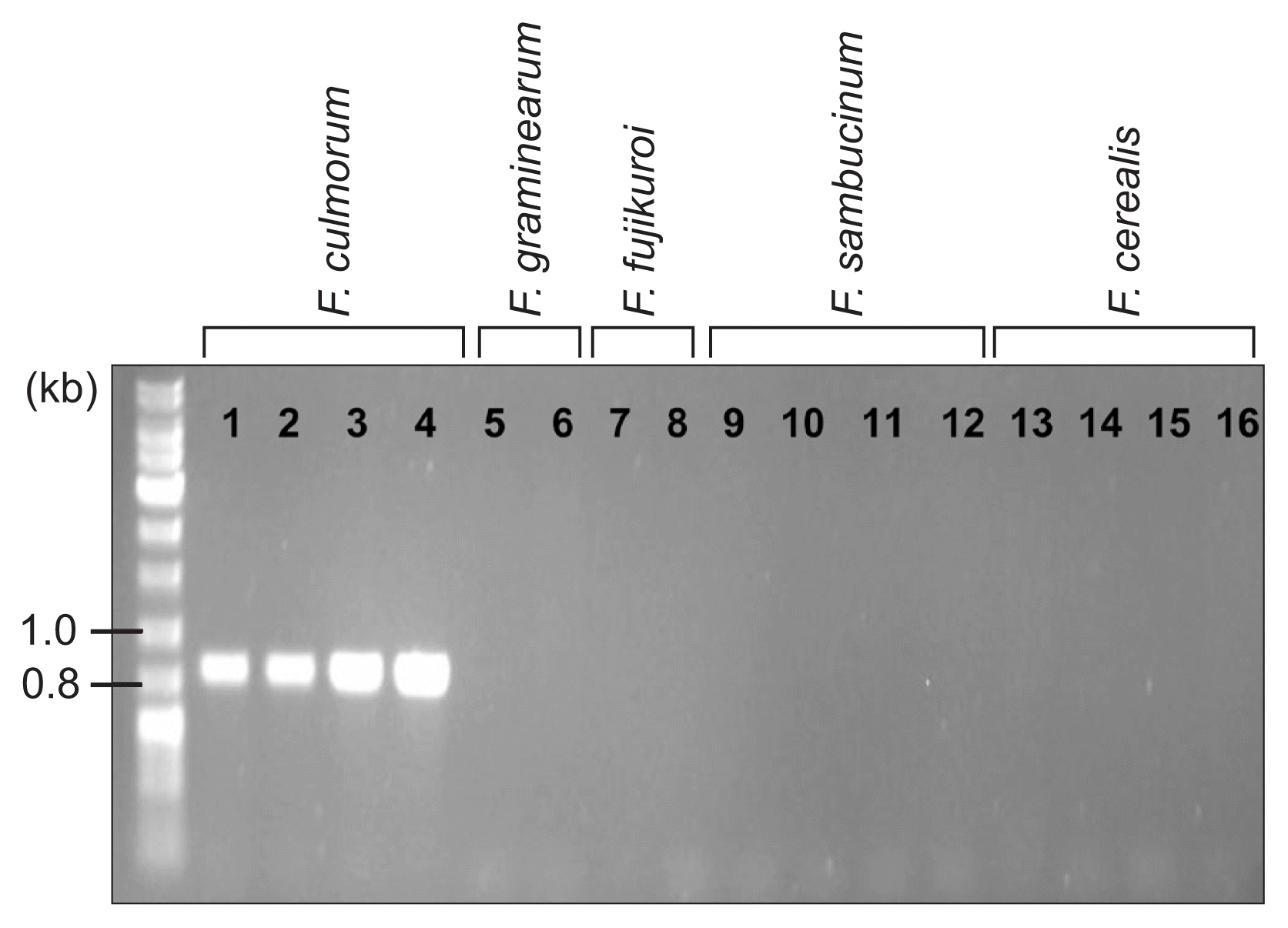
Fig. 6
PCR amplification with mating-type-specific primer sets (for MAT1-1 and MAT1-2). Lanes 1, J1; 2, J2; 3, NRRL14983; 4, NRRL11427; 5, Fusarium graminearum Z3643; 6, F. graminearum PH-1. Note that two F. graminearum strains carried both MAT1-1 and MAT1-2 loci since they are self-fertile (homothallic). Size markers (in kb) are indicated on the left of the gel.

Fig. 7
PCR amplification with trichothecene chemotype-specific primer sets derived from TRI7 and TRI13. Lanes 1, J1; 2, J2; 3, NRRL11427; 4, Fusarium graminearum Z3643; 5, F. asiaticum SCKO4; 6, F. boothii GWS16-4-7; 7, F. vorosii GWR12R1. The size markers (in kb) are indicated on the left of the gel.
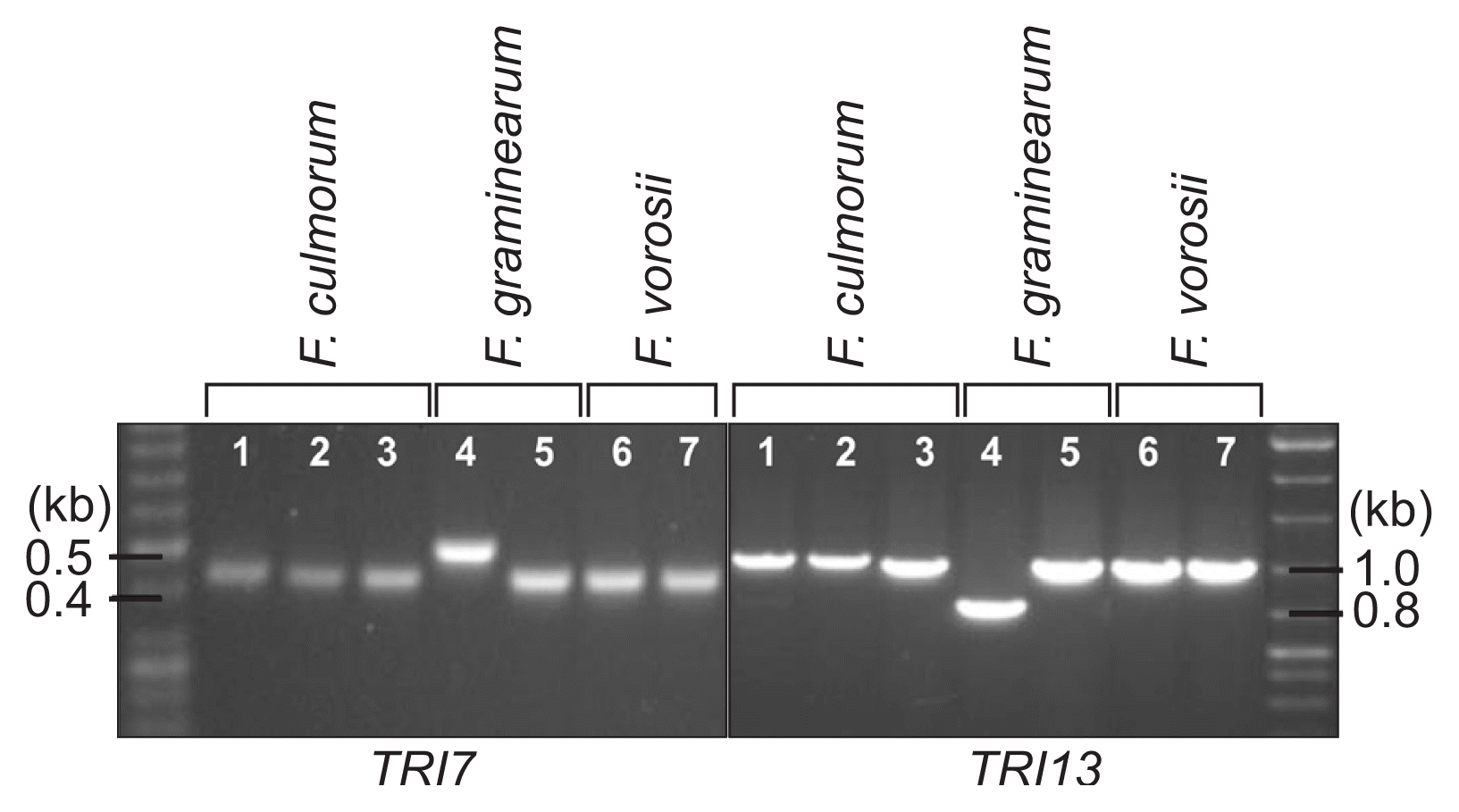
Fig. 8
Development of disease symptoms on wheat heads (A) or in corn stalks (B) when inoculated with Fusarium culmorum J1 and J2 isolates.

Table 1
Primers used in this study
| Target gene | Name | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Phosphate permease (PHO) | PHO1 | ATCTTCTGGCGTGTTATCATG | O’Donnell et al., 2000 |
| PHO6 | GATGTGGTTGTAAGCAAAGCCC | ||
| Histone H3 (H3) | H3F1 | TGGCAAGGCCCCTCGCAAGC | O’Donnell et al., 2000 |
| H3R1 | TTGGACTGGATRGTAACACGC | ||
| Putative reductase (RED) | RED1d | TCTCAGAAAGACGCATATATG | O’Donnell et al., 2000 |
| RED2 | CGTAACTGCGTCATTCGGC | ||
| Ammonia ligase (URA) | URA11 | GAGTATGCCCGCAACGTCATG | O’Donnell et al., 2000 |
| URA16 | AATTATCTCATCGAGACATCC | ||
| Translation elongation factor-1α (TEF1) | EF1 | ATGGGTAAGGA(A/G)GACAAGAC | O’Donnell et al., 2000 |
| EF2 | GGA(G/A)GTACCAGT(G/C)ATCATG | ||
| TRI101 | FculTri101 nF/p1 | ACCCAAAGCCTGTAATTATG | Unpublished |
| FculTri101 nR/p1 | CCAAAGTCGTACTCCCATAA | ||
| TRI7 | Tri7 F | TGCGTGGCAATATCTTCTTCTA | Chandler et al., 2003 |
| Tri7 R | TGTGGAAGCCGCAGA | ||
| TRI13 | Tri13 F | CATCATGAGACTTGTKCRAGTTTGGG | Chandler et al., 2003 |
| Tri13 R | TTGAAAGCTCCAATGTCGTG | ||
| MAT1-1-1 | MAT1-1 F | CTGGAAGAACTGGGCATCGTAA | This study |
| MAT1-1 R | GATATTCTTGTGGCTGGCTACTTT | ||
| MAT1-2-1 | MAT2-1 F | AACACTTACAGCATCATCAGGGCACTCC | This study |
| MAT2-1 R | ATCACCCAGGCACTCGACGTGTGCTTT | ||
| MAT1-2-1 | MAT2-1 F | AACACTTACAGCATCATCAGGGCACTCC | This study |
| MAT2-1 R | ATCACCCAGGCACTCGACGTGTGCTTT |
Table 2
GenBank accession numbers of the genes sequenced in this study
References
Animal, Plant and Fisheries Quarantine and Inspection Agency. 2013. List of plant quarantine fungi in Korea newly revised in 2013. Res Plant Dis. 19:237-241.
Bakan, B, Pinson, L, Cahagnier, B, Melcion, D, Sémon, E and Richard-Molard, D 2001. Toxigenic potential of Fusarium culmorum strains isolated from French wheat. Food Addit Contam. 18:998-1003.


Chandler, EA, Simpson, DR, Chandler, EA, Duncan, RS, Thomsett, MA and Nicholson, P 2003. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterisation of chemotypes of Fusarium graminearum, Fusarium culmorum and Fusarium cerealis. Physiol Mol Plant Pathol. 62:355-367.

Chi, MH, Park, SY and Lee, YH 2009. A quick and safe method for fungal DNA extraction. Plant Pathol J. 25:108-111.

Demeke, T, Clear, RM, Patrick, SK and Gaba, D 2005. Species-specific PCR-based assays for the detection of Fusarium species and a comparison with the whole seed agar plate method and trichothecene analysis. Int J Food Microbiol. 103:271-284.


Gang, G, Miedacjner, T, Schuhmacher, U, Schollenberger, M and Geiger, HH 1998. Deoxynivalenol and nivalenol production by Fusarium culmorum isolates differing in aggressiveness toward winter rye. Phytopathology. 88:879-884.


Goswami, RS and Kistler, HC 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 5:515-525.


Han, YK, Kim, MD, Lee, SH, Yun, SH and Lee, YW 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol Microbiol. 63:768-779.


Hogg, AC, Johnston, RH, Johnston, JA, Klouser, L, Kephart, KD and Dyer, AT 2010. Monitoring fusarium crown rot populations in spring wheat residues using quantitative real-time polymerase chain reaction. Phytopathology. 100:49-57.


Iwama, T, Katsube, K and Ishii, H 2007. Detection of thiophanate- methyl-resistant isolates of Fusarium culmorum, a causal agent of Fusarium head blight on wheat, in Aomori prefecture, northern Japan. Jpn J Phytopathol. 73:162-165.

Jennings, P, Coates, ME, Walsh, K, Turner, JA and Nicholson, P 2004. Determination of deoxynivalenol- and nivalenol- producing chemotypes of Fusarium graminearum isolated from wheat crops in England and Wales. Plant Pathol. 53:643-652.

Jung, B, Lee, S, Ha, J, Park, JC, Han, SS, Hwang, I, Lee, YW and Lee, J 2013. Development of a selective medium for the fungal pathogen Fusarium graminearum using toxoflavin produced by the bacterial pathogen Burkholderia glumae. Plant Pathol J. 29:446-450.



Kerényi, Z, Moretti, A, Waalwijk, C, Oláh, B and Hornok, L 2004. Mating type sequences in asexually reproducing Fusarium species. Appl Environ Microbiol. 70:4419-4423.




Kim, H, Kim, HK, Lee, S and Yun, SH 2015. The white collar complex is involved in sexual development of Fusarium graminearum. PLoS One. 10:e0120293



Kim, JC, Park, AR, Lee, YW, Youn, HJ and Cha, SH 1993. Variation in trichothecene and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Korean J Microbiol. 31:312-317.
Lee, J, Chang, IY, Kim, H, Yun, SH, Leslie, JF and Lee, YW 2009. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl Environ Microbiol. 75:3289-3295.




Lee, T, Oh, DW, Kim, HS, Lee, J, Kim, YH, Yun, SH and Lee, YW 2001. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae by using PCR. Appl Environ Microbiol. 67:2966-2972.




Leslie, JF and Summerell, BA 2006. The Fusarium lab manual. Wiley-Blackwell, Ames, IA, USA.
Miedaner, T, Cumagun, CJR and Chakraborty, S 2008. Population genetics of three important head blight pathogens Fusarium graminearum, F. pseudograminearum and F. culmorum. J Phytopathol. 156:129-139.

Mishra, PK, Fox, RTV and Culham, A 2003. Inter-simple sequence repeat and aggressiveness analyses revealed high genetic diversity, recombination and long-range dispersal in Fusarium culmorum. Ann Appl Biol. 143:291-301.

Muthomi, JW, Schütze, A, Dehne, HW, Mutitu, EW and Oerke, EC 2000. Characterization of Fusarium culmorum isolates by mycotoxin production and aggressiveness to winter wheat. J Plant Dis Protect. 107:113-123.
Nielsen, LK, Jensen, JD, Rodríguez, A, Jørgensen, LN and Justesen, AF 2012. TRI12 based quantitative real-time PCR assays reveal the distribution of trichothecene genotypes of F. graminearum and F. culmorum isolates in Danish small grain cereals. Int J Food Microbiol. 157:384-392.


Obanor, F, Erginbas-Orakci, G, Tunali, B, Nicol, JM and Chakraborty, S 2010. Fusarium culmorum is a single phylogenetic species based on multilocus sequence analysis. Fungal Biol. 114:753-765.


O’Donnell, K, Kistler, HC, Tacke, BK and Casper, HH 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci U S A. 97:7905-7910.



Parry, DW, Jenkinson, P and McLeod, L 1995. Fusarium ear blight (scab) in small grain cereals-a review. Plant Pathol. 44:207-238.

Pasquali, M, Giraud, F, Brochot, C, Cocco, E, Hoffmann, L and Bohn, T 2010. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. Int J Food Microbiol. 137:246-253.


Scherm, B, Balmas, V, Spanu, F, Pani, G, Delogu, G, Pasquali, M and Migheli, Q 2013. Fusarium culmorum: causal agent of foot and root rot and head blight on wheat. Mol Plant Pathol. 14:323-341.


Stepień, Ł, Popiel, D, Koczyk, G and Chełkowski, J 2008. Wheat-infecting Fusarium species in Poland--their chemotypes and frequencies revealed by PCR assay. J Appl Genet. 49:433-441.



Thompson, JD, Higgins, DG and Gibson, TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680.



Waalwijk, C, Kastelein, P, Vries, I, Kerényi, Z, Van der Lee, T, Hesselink, T, Köhl, J and Kema, G 2003. Major changes in Fusarium spp. in wheat in the Netherlands. Eur J Plant Pathol. 109:743-754.

Wagacha, JM and Muthomi, JW 2007. Fusarium culmorum: infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Protect. 26:877-885.

West, JS, Holdgate, S, Townsend, JA, Edwards, SG, Jennings, P and Fitt, BDL 2012. Impacts of changing climate and agronomic factors on fusarium ear blight of wheat in the UK. Fungal Ecol. 5:53-61.

Yli-Mattila, T 2010. Ecology and evolution of toxigenic Fusarium species in cereals in northern europe and Asia. J Plant Pathol. 92:7-18.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




