 |
 |
| Plant Pathol J > Volume 33(4); 2017 > Article |
Abstract
Chrysanthemums (Chrysanthemum morifolium) are susceptible to tobacco mosaic virus (TMV). TMV-based expression vectors have been used in high-throughput experiments for production of foreign protein in plants and also expressing green fluorescent protein (GFP) to allow visualization of TMV movement. Here, we used TMV expressing the GFP to examine the infection of chrysanthemum by a TMV-based expression vector. Viral replication, movement and GFP expression by TMV-GFP were verified in upper leaves of chrysanthemums up to 73 days post inoculation (dpi) by RT-PCR. Neither wild-type TMV nor TMV-GFP induced symptoms. GFP fluorescence was seen in the larger veins of the inoculated leaf, in the stem above the inoculation site and in petioles of upper leaves, although there was no consistent detection of GFP fluorescence in the lamina of upper leaves under UV. Thus, a TMV-based expression vector can infect chrysanthemum and can be used for the in vivo study of gene functions.
Chrysanthemums (Chrysanthemum morifolium) are one of the most popular and important cut flowers in the world. The international market for cut and potted chrysanthemums has greatly expanded and chrysanthemums are commercially very important for the floral industry (Liu and Gao, 2007; Tomassoli et al., 2004). Chrysanthemum is one of the three most economically valuable cut flowers in Korea (MAFRA, 2014).
To date, chrysanthemum is reported to be susceptible to nine viruses and two viroids, some of which cause serious disease and affect chrysanthemums commercially (Verma et al., 2003). Chrysanthemum plants can be infected by viruses including chrysanthemum virus B, cucumber mosaic virus, tomato aspermy virus, tomato spotted wilt virus (Bouwen and Zaayen, 1995), potato virus X (Navalinskiene and Samuitiene, 1996), potato virus Y, tobacco mosaic virus (TMV) (Zhao et al., 2015) and turnip mosaic virus (Chen et al., 1999, 2000). Beside these viruses, two viroids, chrysanthemum stunt viroid (Diener and Lawson, 1973) and chrysanthemum chlorotic mottle viroid infect chrysanthemum plants (Flores et al., 2003).
TMV has even been reported to infect chrysanthemum, but no description of the pathology has been given (Liu et al., 2014; Zhao et al., 2015). In other hosts, the use of TMV-based vectors for the transient expression of foreign genes in plants has attracted considerable interest (Scholthof et al., 1996). TMV-based vectors expressing the green fluorescent protein (GFP) have been used to follow virus movement (Canto and Palukaitis, 2002; Casper and Holt, 1996; Shivprasad et al., 1999). The vector TMV-GFP 1056, which is derived from TMV 30B-GFP (Shivprasad et al., 1999) with modification in the movement protein (MP) gene, spread systemically more efficiently than earlier derived vectors (Canto and Palukaitis, 2002). Here, we studied the infection process of TMV-GFP 1056 in chrysanthemum, to determine whether a TMV-based expression vector could be used in chrysanthemum and how infection of chrysanthemum by TMVGFP 1056 compared to infection by wild-type (WT) TMV.
C. morifolium cv. ‘Vivid Scarlet’ was grown from cuttings of virus- and viroid-free mother plants. All plants were grown in a growth room at 23°C with a 16-h photoperiod. Chrysanthemums, with six leaves to eight leaves after cutting and rooting, were used for infection with WT TMV and a TMV-based vector. These were inoculated mechanically on two leaves. First, the stem was slashed with a razor blade and then inoculum was rubbed onto Carborundum-dusted leaves and the cut stem, to improve the chance of infection. The inoculum was an extract from infected Nicotiana benthamiana plants, previously inoculated with either an extract from a WT TMV-infected dried leaf or RNA transcripts of TMV-GFP 1056, as described previously (Canto and Palukaitis, 2002). To observe GFP-derived fluorescence, virus-inoculated plants were viewed under a UV lamp (Mag-Lite; Mag Instrument, Ontario, CA, USA) and photographed using a camera equipped with a screw-in filter (Yellow 12; Tiffen, Hauppauge, NY, USA) to reduce red autofluorescence from the chloroplasts.
Leaves from mock and virus-inoculated plants at 18, 30, and 73 days post inoculation (dpi) were ground in liquid nitrogen using a mortar and pestle, and total RNAs were extracted with an IQeasy™ Extraction Mini Kit (iNtRON Biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. RNA concentrations were measured using the Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
Complementary DNAs (cDNAs) were synthesized from extracted RNA, using the RevertAid Reverse transcriptase (Thermo Fisher Scientific), following the manufacturer’s instructions. For PCR amplification, TMV gene-specific primers, designed based on the sequence of TMV RNA, and GFP-specific primers were used (Table 1). TaKaRa Ex Taq (Takara, Tokyo, Japan) polymerase was used with the synthesized cDNA to perform PCR amplification, according to the manufacturer’s instructions. PCR products were analyzed by electrophoresis in 1.5% agarose gels followed by ethidium bromide staining and were observed under UV light.
Infection of chrysanthemum by WT TMV did not cause any stunting or foliar symptoms on either the inoculated leaves or upper leaves (data not shown). Similarly, TMVGFP infected chrysanthemums showed no symptoms on the leaves under visible light at 18 dpi (Fig. 1A, 2A, B; upper images) or at 30 dpi (Fig. 3). To confirm replication and systemic infection of WT TMV and TMV-GFP, total RNAs were extracted from inoculated and upper leaves, and the TMV MP gene was amplified by RT-PCR using specific primers. The target band from RT-PCR was detected in both the inoculated leaf and upper leaves for both WT TMV and TMV-GFP (Fig. 1C). TMV-GFP-infected chrysanthemums showed fluorescence on occasional veins in inoculated leaves under UV light at ~18 dpi and fluorescence also was seen in the petiole of upper leaves (Fig. 1A, 2A, B), but not over the surface of the leaf lamina versus in N. benthamiana, which showed systemic green fluorescence early after inoculation (Fig. 1B). This was also the case for upper leaves in chrysanthemum plants infected with TMV-GFP at 30 dpi (Fig. 3) and at 73 dpi (images not shown). At 30 dpi, green fluorescence also could be seen in the stem (Fig. 3C).
To confirm TMV spread and the stability of the GFP insert in the TMV-GFP in upper leaves, the presence of the various genes of TMV-GFP (RNA-dependent RNA polymerase, MP, coat protein, and GFP) was assessed by RT-PCR at 30 dpi and 73 dpi. All tested leaves showed the presence of TMV-specific genes and the GFP gene (Fig. 4A, B). The upper leaf at 73 dpi showed the same amplification profile as the upper leaf at 30 dpi from the same plant (Fig. 4B vs 4A). Thus, TMV-GFP was infectious in chrysanthemum and retained the GFP insert for over 10 weeks, but showed a delayed systemic infection with no visible symptoms, compared with N. benthamiana (Fig. 1,-3).
In conclusion, inoculation of chrysanthemum with either WT TMV or the expression vector TMV-GFP 1056 resulted in a symptomless infection. TMV-GFP infected C. moriforium cv. ‘Vivid Scarlet’ systemically with limited green fluorescence under UV light, unlike N. benthamiana which shows stronger fluorescence. A reduced rate of spread of TMV-GFP versus TMV is often the case with viral vectors and has been noted before with TMV-GFP in both N. benthamiana and tobacco (Canto and Palukaitis, 2002; Rabindran and Dawson, 2001). Our results are comparable with using TRV-GFP, which gave systemic infection in chrysanthemum but GFP fluorescence was seen only in the stems after agro-inoculation (Tian et al., 2014). Nevertheless, using RT-PCR, we confirmed the spread of TMV, by the presence of the TMV genes and the GFP insert from TMV-GFP infection in upper leaves at 30 dpi and 73 dpi. Hence, TMV-GFP 1056 can be expected to be useful as an expression vector for functional analysis of specific genes or proteins in chrysanthemum plants.
Acknowledgments
This study was supported by grant number PJ011309032017 from the Next Generation BioGreen21 Program of the Rural Development Administration, Republic of Korea.
Fig. 1
Infection by TMV-GFP in chrysanthemum and in Nicotiana benthamiana plants. (A) Symptomless infection induced by TMV-GFP under visible light (Vis; upper panel) or UV light (UV; lower panel). (B) Systemic stunting, leaf curl and epinasty induced by TMV-GFP in N. benthamiana under visible light (upper panel) or UV light (lower panel). (C) Stained agarose gel of RT-PCR products of the TMV movement protein (MP) gene, using RNA extracted from wild-type TMV or TMV-GFP infected chrysanthemum leaves at 18 days post inoculation, either the inoculated leaf (Inoc), or an upper systemically infected leaf (UpL). M, 1 kb + DNA ladder marker (Invitrogen, USA). PC, positive control (RT-PCR product of the TMV MP gene derived from TMV RNA). Asterisk, expected position of the MP gene RT-PCR product. TMV, tobacco mosaic virus; GFP, green fluorescent protein.
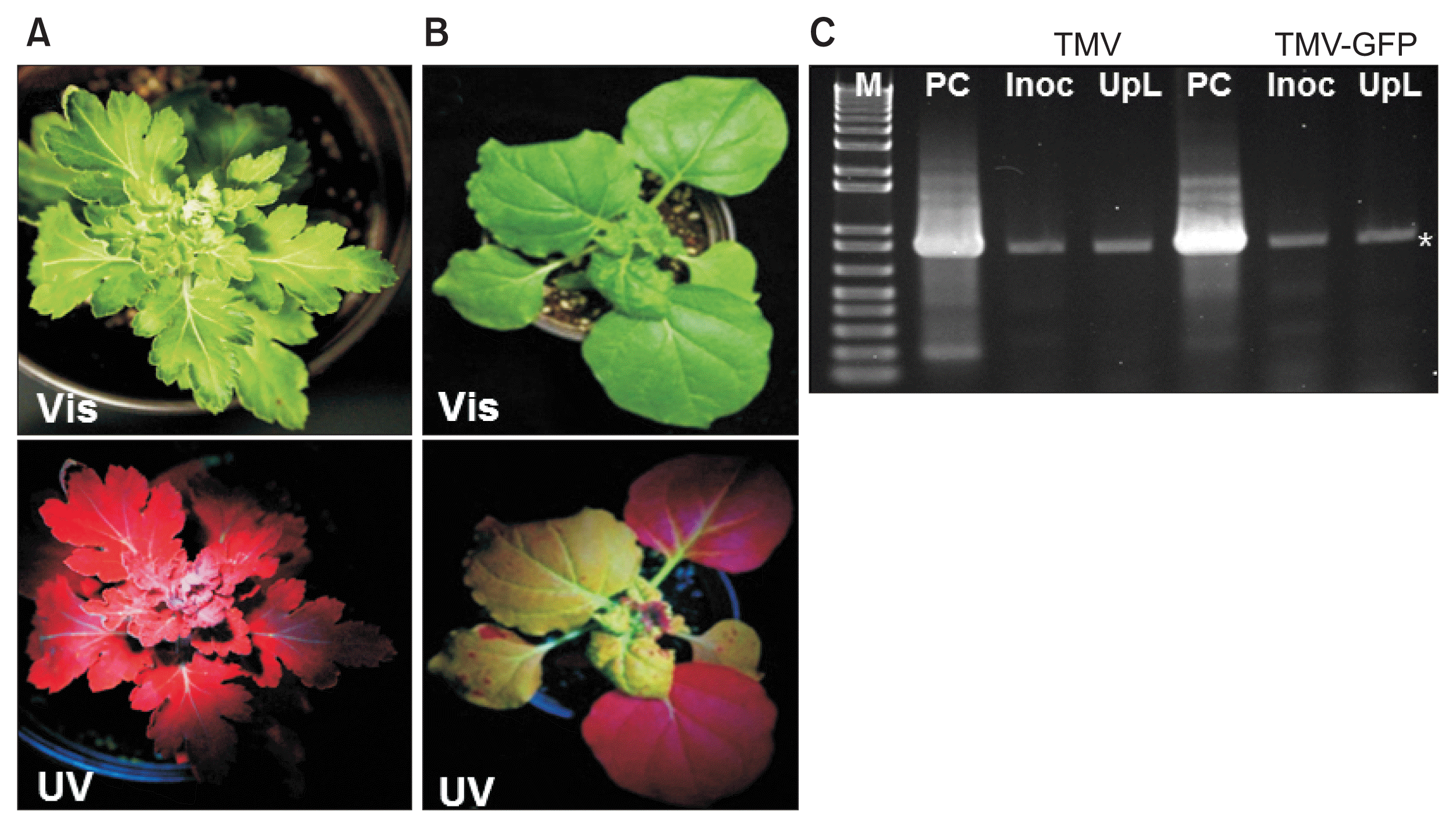
Fig. 2
GFP fluorescence detection in TMV-GFP infected chrysanthemum leaves at 18 days post inoculation. Expression of green fluorescence under visible light (Vis; upper panels) and UV light (UV; lower panels). (A) Inoculated (Inoc.) leaf. (B) Upper leaf of inoculated plant. Arrows point out fluorescence in major veins in (A) and in the petiole and stem in (B). (C) Leaf of a non-inoculated plant. TMV, tobacco mosaic virus; GFP, green fluorescent protein.
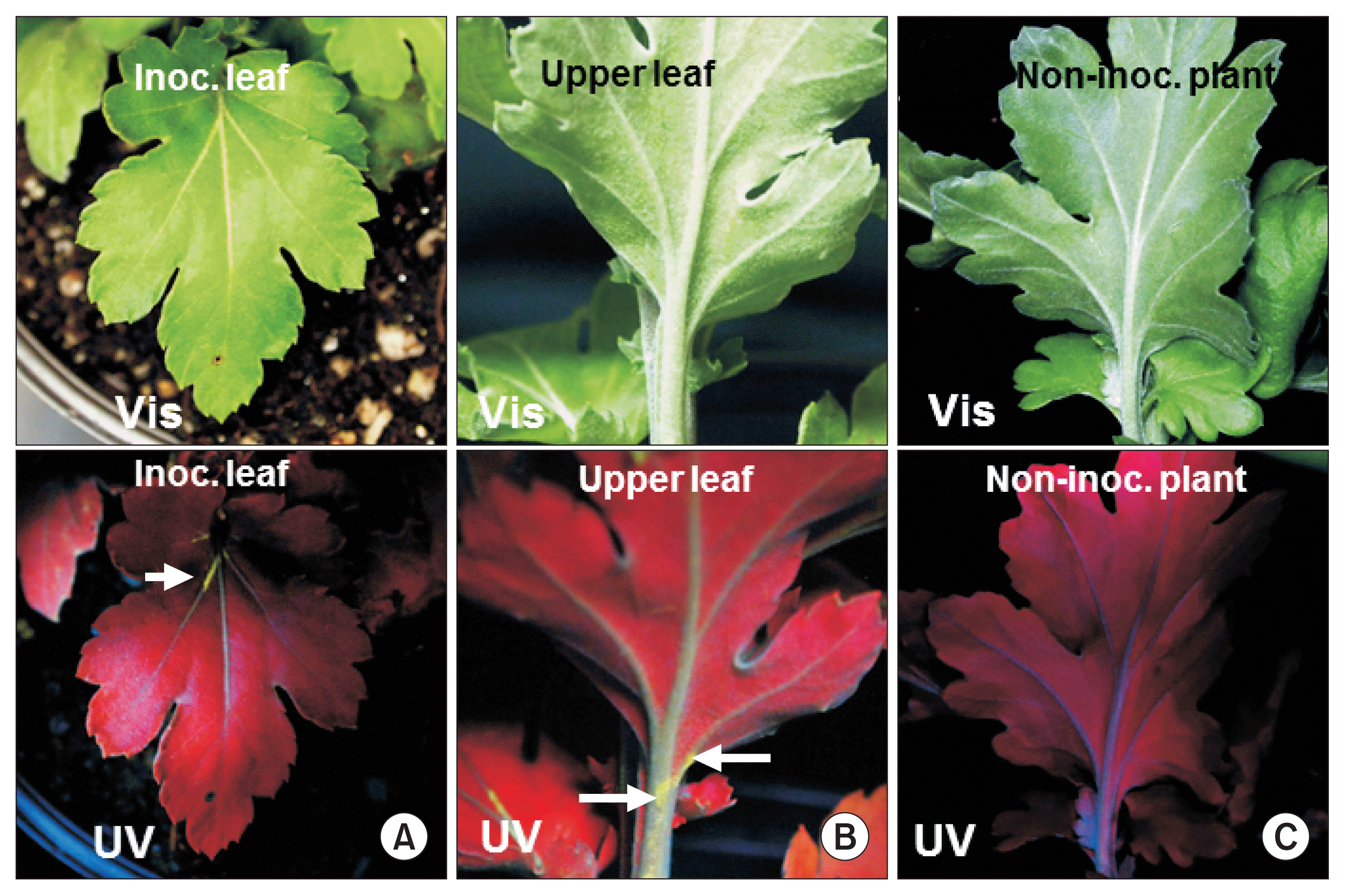
Fig. 3
GFP fluorescence detection in TMV-GFP infected chrysanthemum leaves at 30 days post inoculation. (A) TMV-GFP inoculated plant, showing identifying leaves that were inoculated (Inoc.), or two upper leaves (UpL1 and UpL2) examined in close-up in (B) and (C), under visible light (Vis; left panels) or UV light (UV; right panels). (B) Upper (adaxial) surface of inoculated leaf, first upper leaf and second upper leaf. (C) Lower (abaxial) surface of inoculated leaf and first upper leaf. Arrows point out fluorescence in major veins in (B) and in the petiole and stem in (C). TMV, tobacco mosaic virus; GFP, green fluorescent protein.
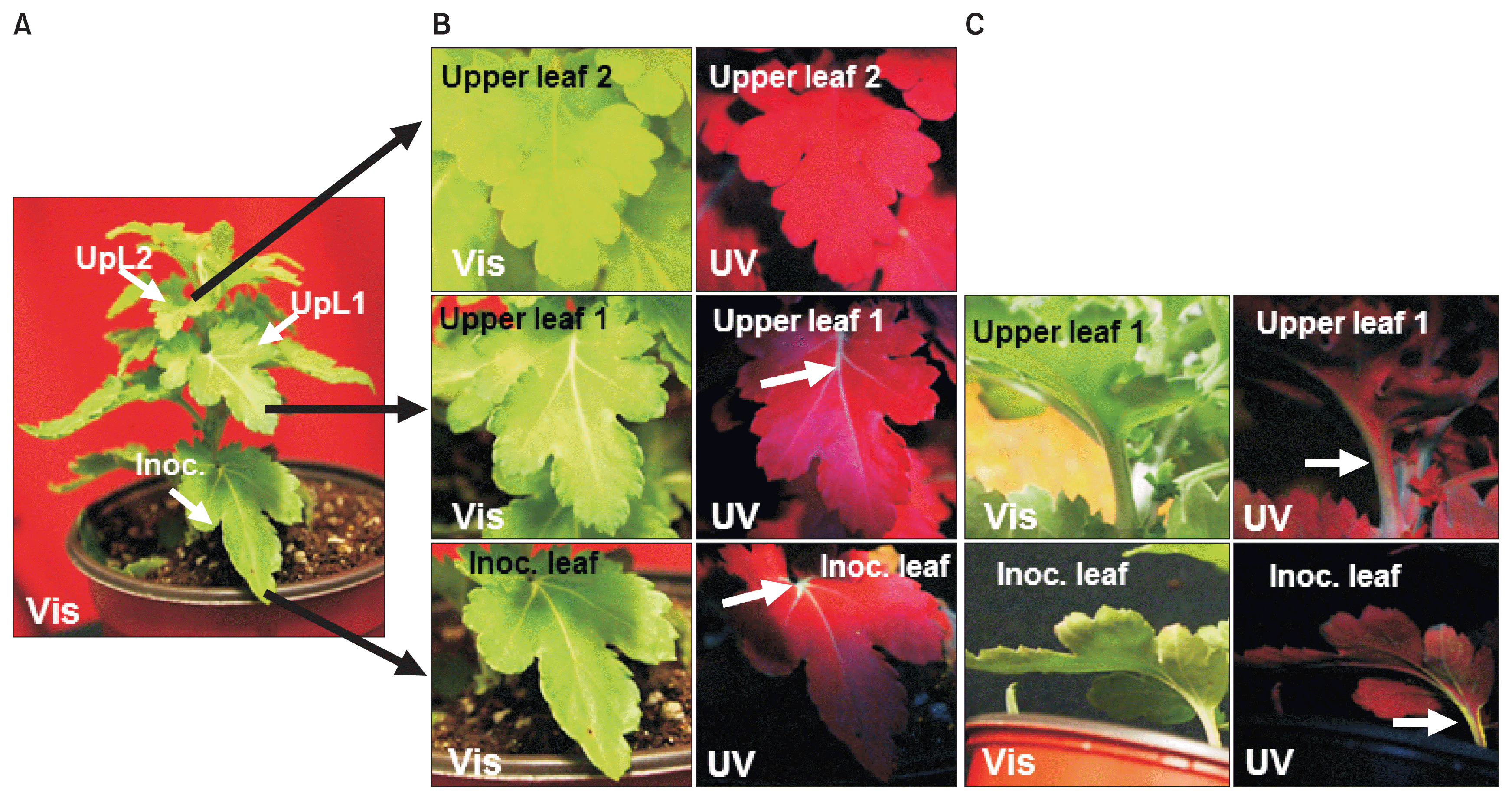
Fig. 4
Detection of TMV genes and GFP sequences in chrysanthemum plants infected for 30 and 73 days post inoculation (dpi). (A) RT-PCR products using RNA extracted at 30 dpi from an inoculated leaf (Inoc) or upper leaves (UpL1 and UpL2) of a TMV-GFP infected plant, with primers corresponding to the movement protein gene (MP), the coat protein gene (CP) or the GFP gene (GFP). (B) The RT-PCR products obtained using RNA extracted from either an upper infected leaf (UpL3) at 73 dpi, or from a mock-inoculated leaf (Mock; NC, negative control), using the same primers as in (A), but also primers for the RNA-dependent RNA polymerase gene (RdRp). (C) Positive control (PC). The RT-PCR products obtained using RNA extracted at 18 dpi from an Nicotiana benthamiana infected with TMV-GFP. M, 1 kb + DNA ladder marker (Invitrogen, USA). TMV, tobacco mosaic virus; GFP, green fluorescent protein.
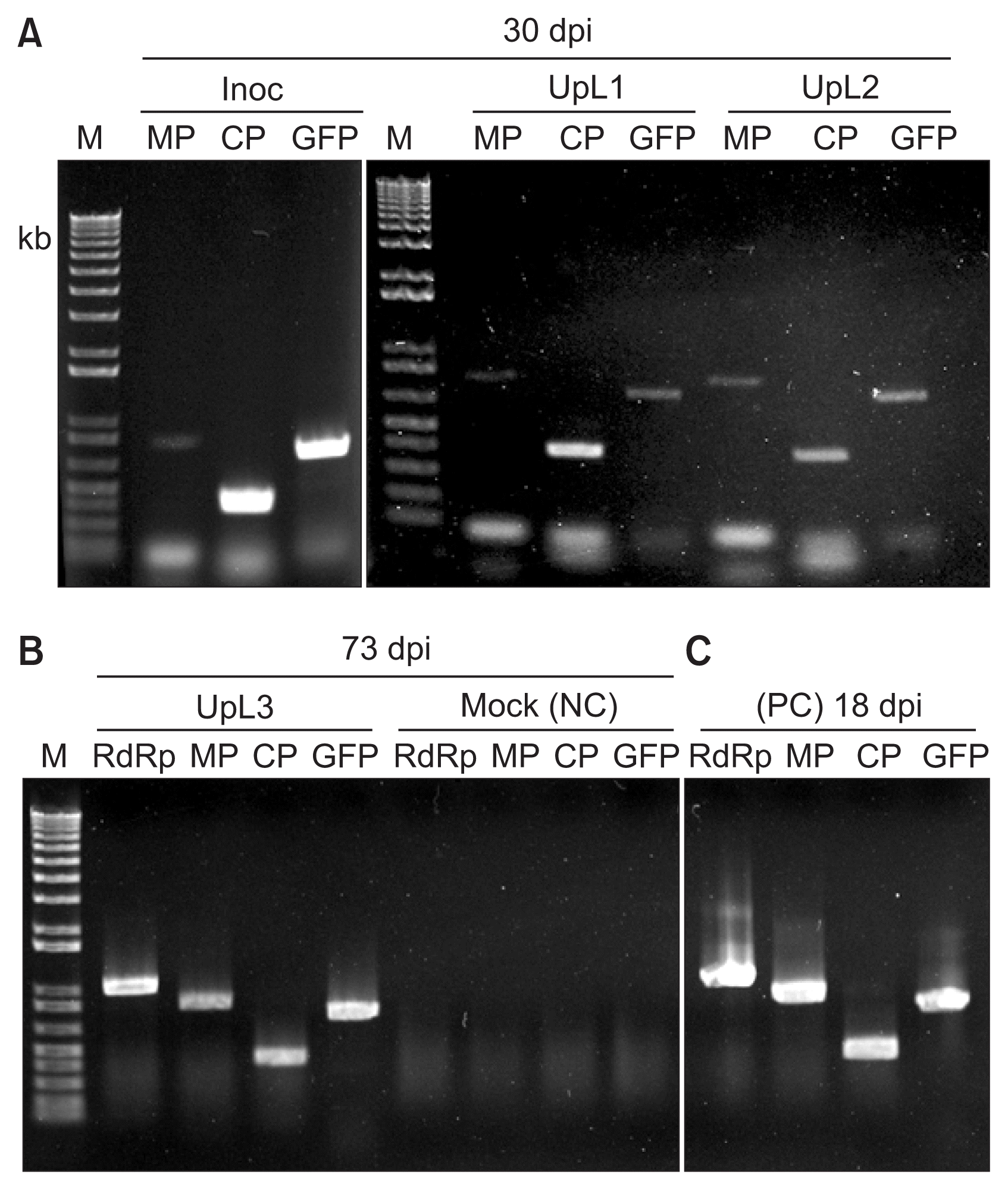
Table 1
Specific primer sets for a detection of TMV and TMV-GFP
References
Bouwen, I and Zaayen, AV 1995. Chrysanthemum. In: Virus and virus-like diseases of bulb and flower crops, eds. by G Loebenstein, RH Lawson and AA Brunt, 396-408. John Wiley & Sons, Chichester, UK.
Canto, T and Palukaitis, P 2002. Novel N gene-associated, temperature-independent resistance to the movement of tobacco mosaic virus vectors neutralized by a Cucumber mosaic virus RNA1 transgene. J Virol. 76:12908-12916.




Casper, SJ and Holt, CA 1996. Expression of the green fluorescent protein-encoding gene from a tobacco mosaic virus-based vector. Gene. 173:69-73.


Chen, TM, Chen, MJ and Yeh, SD 2000. Characterization of the coat protein gene of a turnip mosaic virus isolate infecting garland chrysanthemum. Plant Prot Bull Taipei. 42:83-96.
Chen, TM, Chen, YK, Chen, MJ and Yeh, SD 1999. Host reactions, cytological characteristics and serological properties of a Turnip mosaic virus isolate causing albinic mosaic disease of garland chrysanthemum. Plant Pathol Bull. 8:73-82.
Flores, R, de la Peña, M and Navarro, B 2003. Chrysanthemum chlorotic mottle viroid. In: Viroids, eds. by A Hadidi, R Flores, JW Randles and JS Semancik, 224-227. CSIRO Publishing, Collingwood, Australia.

Liu, XL, Zhao, XT, Muhammad, I, Ge, BB and Hong, B 2014. Multiplex reverse transcription loop-mediated isothermal amplification for the simultaneous detection of CVB and CSVd in chrysanthemum. J Virol Methods. 210:26-31.


Liu, Z and Gao, S 2007. Micropropagation and induction of autotetraploid plants of Chrysanthemum cinerariifolium (Trev.) Vis. In Vitro Cell Dev Biol: Plant. 43:404-408.


[MAFRA] Ministry of Agriculture, Food and Rural Affairs. 2014 onwards Status of horticulture in Korea, 2013 (in Korean). URL http://library.mafra.go.kr/skyblueimage/14503.pdf. 12 July 2017.
Navalinskiene, M and Samuitiene, M 1996. Viral diseases of flower plants. 9. Results of identification of viruses affecting chrysanthemum. Biologija. 4:66-74.
Rabindran, S and Dawson, WO 2001. Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology. 284:182-189.


Scholthof, HB, Scholthof, KB and Jackson, AO 1996. Plant virus gene vectors for transient expression of foreign proteins in plants. Annu Rev Phytopathol. 34:299-323.


Shivprasad, S, Pogue, GP, Lewandowski, DJ, Hidalgo, J, Donson, J, Grill, LK and Dawson, WO 1999. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology. 255:312-323.


Tian, J, Pei, H, Zhang, S, Chen, J, Chen, W, Yang, R, Meng, Y, You, J, Gao, J and Ma, N 2014. TRV-GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J Exp Bot. 65:311-322.


Tomassoli, L, Faggioli, F, Zaccaria, A, Caccia, R, Albani, M and Barba, M 2004. Molecular diagnosis of Chrysanthemum stunt viroid for routine indexing. Phytopathol Mediterr. 43:285-288.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




